Paper2Agent proposes a bold shift in how scientific results move from publication to practice, by converting a research paper and its public code into a conversational AI agent that can run the paper’s methods, answer questions, reproduce figures, and apply workflows to new data. This work by Jiacheng Miao et al centers the Model Context Protocol (MCP) as the standard interface between agentic LLMs and scientific tools.
Grounded in a multi-agent pipeline and validated on case studies in genomics and single-cell biology, the authors report that Paper2Agent generates robust, reusable MCP tools from runnable tutorials, packages them as a server, and connects them to chat-based developer assistants like Claude Code (Anthropic, 2025). This article unpacks the system’s architecture, reviews its reported results, compares it with adjacent efforts like Code2MCP (Ouyang et al., 2025) and MCP for Science & HPC (Pan et al., 2025), and highlights practical trade offs for adoption.
Explore our overview of the Paper2Agent Github Repository here
Key Takeaways
- Automated paper-to-agent conversion: Paper2Agent derives MCP servers directly from a paper’s codebase using tutorial scanning, execution, tool extraction, and testing.
- MCP abstraction: Each “paper MCP” exposes three pillars — tools, resources, and prompts — enabling agents to run methods, retrieve assets, and follow encoded workflows.
- Case-study validation: Reported 100% accuracy on curated and novel AlphaGenome queries plus reproduction of human-executed results for TISSUE and Scanpy workflows; AlphaGenome details and API in (Avsec et al., 2025).
- Developer ergonomics: Claude Code integration makes the MCP servers usable in day-to-day coding sessions while hosting on Hugging Face Spaces avoids local dependency issues.
- Performance profile: Authors note typical end-to-end runtime of roughly 30 minutes to several hours depending on repository complexity and compute, with one-time costs for complex repos documented in the project README.
- Ecoystem fit: Complements efforts like Code2MCP to reduce the (N × M) integration problem by standardizing tool access over MCP (Ouyang et al., 2025).
Inside The Approach
Paper2Agent represents a paper as a remotely deployable MCP server and then connects that server to a chat agent. The Model Content Protocol is an open standard for exposing data, tools, and workflows to LLM-based applications developed and supported by Anthropic.
The conversion produces three components:
(i) MCP Tools — executable functions wrapping the paper’s core methods;
(ii) MCP Resources — structured links to manuscripts, code, datasets, and figures;
(iii) MCP Prompts — concise, paper-derived workflow templates that guide the agent through multi-step analyses.
A multi-agent pipeline performs the heavy lifting. where an environment manager provisions an isolated, pinned Python setup then a tutorial scanner discovers runnable notebooks and step-by-step guides worth distilling. Next a tutorial executor runs them in an auditable way to capture inputs, outputs, and implicit assumptions. Followed by a tool-extractor that implements parameterized functions which generalize beyond the examples. Finally, a test-verifier-improver loop generates tests, runs them, diagnoses failures, and iteratively hardens the tools before the MCP server is assembled (Miao et al., 2025).
The MCP server can be launched locally or hosted remotely (e.g., in a Hugging Face Space) and then attached to a coding assistant. The project’s README documents typical usage, including repository targeting, tutorial filtering, and optional API keys for methods like AlphaGenome. See the GitHub project for current scripts and parameterization (Miao et al., 2025).
# Example: Process only specific tutorials by title or URL on github:
bash Paper2Agent.sh \
--project_dir <PROJECT_DIR> \
--github_url https://github.com/scverse/scanpy \
--tutorials "Preprocessing and clustering" Why It Matters
Reproducibility and adoption are persistent bottlenecks in many disciplines. Even when code is available, reproducing figures or applying methods often demands environment wrangling, undocmented parameters, and trial-and-error.
Paper2Agent reframes this gap and it turns the paper into an interface that is both conversational and executable. If a method is packaged as tools, resources, and prompts and those tools are tested against the paper’s own examples, then an agent can help re-run, explain, and adapt the method for new data.
At the ecosystem level, MCP reduces coupling between agents and services. Code2MCP targets similar friction: the N × M problem in which many models must integrate with many tools, by automating the wrapping of GitHub repos as MCP services (Ouyang et al., 2025).
Meanwhile, experiences from science cyberinfrastructure show that thin MCP servers over mature services like data transfer and compute make agent access more uniform in HPC and lab settings (Pan et al., 2025). Paper2Agent fits squarely into this trajectory by focusing on research papers and their tutorials.
Results, Figures, And Technical Nuances
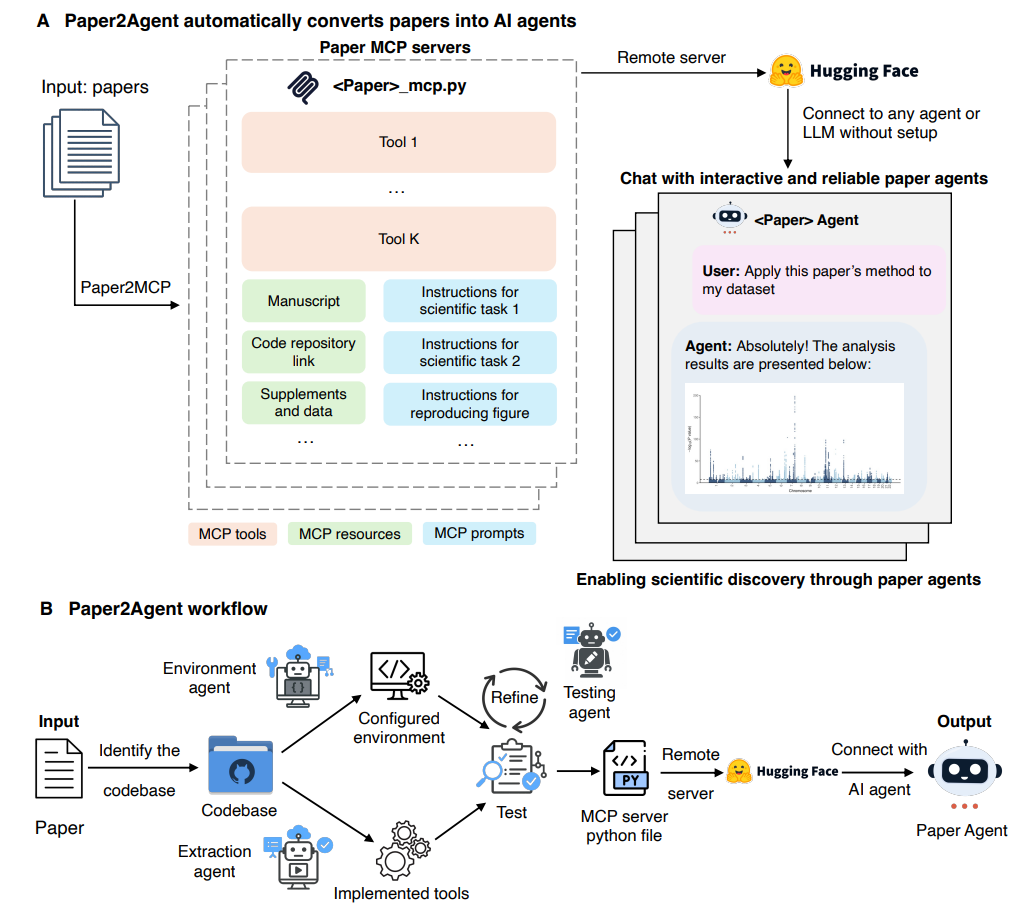
Figure 1: Overview of the Paper2Agent. (A) Paper2Agent turns research papers into interactive AI agents by building remote MCP servers with tools, resources, and prompts. Connecting an AI agent to the server creates a paper-specific agent for diverse tasks. (B) Workflow of Paper2Agent. It starts with codebase extraction and automated environment setup for reproducibility. Core analytical features are wrapped as MCP tools, then validated through iterative testing. The resulting MCP server is deployed remotely and integrated with an AI agent, enabling natural-language interaction with the paper’s methods and analyses.
Figure 1 in the paper sketches the end-to-end flow: from codebase identification and environment setup to tool extraction, testing, and MCP server deployment. The key is that validated functionality moves from the tutorial into parameterized tools, with inputs and outputs documented for agent use.

AlphaGenome case study — Figure 2. Paper2Agent reports generating 22 MCP tools in roughly 3 hours on a personal laptop. Tools include single and batch variant scoring across modalities and visualization utilities. The paper benchmarks an AlphaGenome agent on curated tutorial queries and a set of novel prompts, reporting 100% accuracy on numerical answers for both groups.
The authors also demonstrate automated, step-wise interpretation of a GWAS locus by planning variant scoring, filtering trait-relevant tissues, rendering modality tracks, and assembling a report. Notably, their re-analysis highlights SORT1 as a likely causal gene for the LDL locus alongside CELSR2 and PSRC1, with supporting GTEx eQTL evidence discussed in the text (Miao et al., 2025); background on AlphaGenome’s API and modalities is available in the official repo (Avsec et al., 2025) and eQTL context from GTEx (GTEx Consortium, 2020).
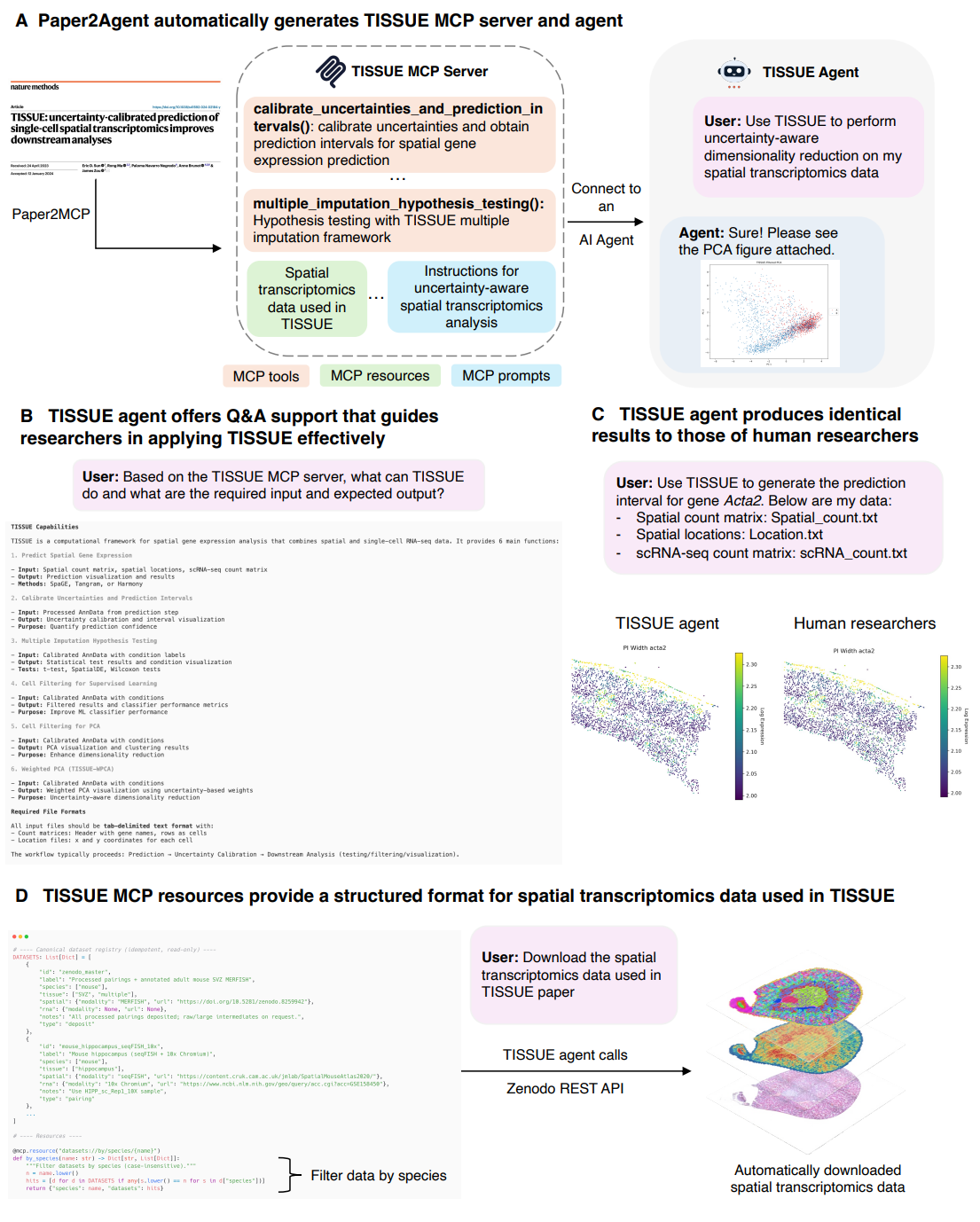
TISSUE case study — Figure 3. The TISSUE agent centers uncertainty-aware spatial transcriptomics. Paper2Agent reports six tools spanning prediction, prediction intervals, and downstream analyses like hypothesis testing and PCA with uncertainty weighting. The agent also answers method questions and exposes an MCP resource registry that links to datasets with standardized metadata, enabling automated downloads through APIs like Zenodo. The paper shows matching results between the agent and human-executed pipelines on a mouse cortex dataset (Miao et al., 2025); the underlying TISSUE method is described in (Sun et al., 2024).
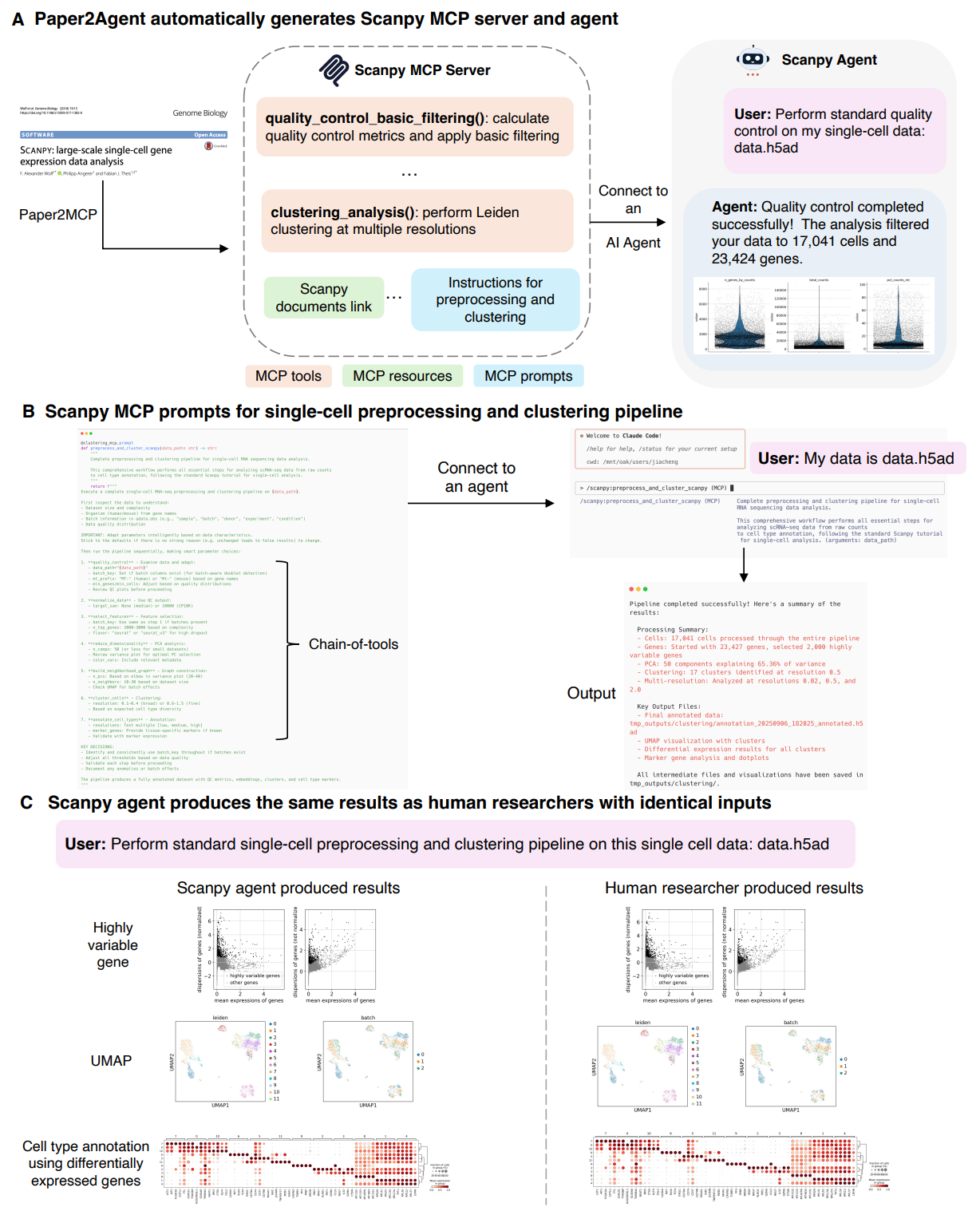
Scanpy case study — Figure 4. The Scanpy agent focuses on a standard preprocessing and clustering pipeline for single-cell RNA-seq. Paper2Agent reports seven tools produced in about 45 minutes, plus MCP prompts that encode the canonical order of operations — QC, normalization, feature selection, dimensionality reduction, graph construction, clustering, and basic annotation. The authors demonstrate reproducing human researcher results on three 10x Genomics PBMC datasets not in the Scanpy codebase. For Scanpy background and reference materials, see (Wolf et al., 2018).
Deep technical notes.
The environment isolation step is a guardrail for executing third-party tutorials that assume specific versions of NumPy, SciPy, or visualization libraries. While the test-verifier-improver loop prunes unreliable tools by removing MCP decorators when failures persist. Tools keep traceable links back to source code to support audit and debugging. And because MCP is transport-agnostic, the same server can be attached to multiple clients in parallel, from Claude Code to other MCP-capable IDE agents, easing team workflows.
Operational considerations.
Some targets require credentials or paid APIs. AlphaGenome, for example, uses an API key with specific terms for non-commercial use. The Paper2Agent README also notes time and cost expectations when using managed LLMs.
Finally, not every codebase is equally amenable to agentification; missing data, brittle notebooks, or implicit assumptions can limit what can be safely exposed. The authors explicitly frame this as a virtue: the ease of converting a paper into an agent becomes a practical measure of reproducibility (Miao et al., 2025).
Conclusion
Paper2Agent introduces a useful abstraction for closing the loop between publication and practice. By extracting tested tools from tutorials, structuring resources, and encoding workflows as prompts, a paper becomes an interactive artifact. The reported results on AlphaGenome, TISSUE, and Scanpy suggest that for well-maintained repos, this approach can reproduce results and generalize to new queries.
Near term, this is valuable for method authors who want their work to be used reliably, and for practitioners who want a lower-friction onramp to modern computational biology and related domains.
Limitations remain: agent quality tracks codebase quality, and thorough evaluation still requires domain expertise. Longer term, expect composite agents that span related papers, plus journals and conferences that standardize authoring patterns to ease agentification. Readers can explore the project code and public MCP servers to try the approach on their own data (Miao et al., 2025).
Definitions
Model Context Protocol (MCP): An open standard for exposing data, tools, and prompts to AI applications in a uniform way, allowing agents to discover and invoke capabilities consistently (Hou et al., 2025).
MCP Tool: An executable function with typed inputs-outputs that wraps a unit of method logic from the paper, designed for agent invocation.
MCP Resource: A standardized pointer or asset bundle that makes manuscripts, datasets, figures, and code discoverable to the agent.
MCP Prompt: A concise, structured instruction template that encodes a multi-step workflow derived from the paper’s text or tutorials.
AlphaGenome: A DeepMind framework and API for multimodal predictions over DNA sequences, including variant effect prediction across modalities (Avsec et al., 2025).
TISSUE: A method for uncertainty-calibrated spatial transcriptomics analysis, including prediction intervals and downstream use of uncertainty (Sun et al., 2024).
Scanpy: A Python toolkit for single-cell RNA-seq analysis used widely for preprocessing, clustering, visualization, and more (Wolf et al., 2018).
GWAS: Genome-wide association study linking genetic variants to traits or diseases; often analyzed alongside eQTL resources like GTEx (GTEx Consortium, 2020).
References
(Miao et al., 2025) Paper2Agent: Reimagining Research Papers As Interactive and Reliable AI Agents.
(Miao et al., 2025) Paper2Agent repository.
(Avsec et al., 2025) AlphaGenome API and documentation.
(Wolf et al., 2018) Scanpy: large-scale single-cell gene expression data analysis.
(Sun et al., 2024) TISSUE: uncertainty-calibrated prediction in spatial transcriptomics.
(Hou et al., 2025) Model Context Protocol: landscape and security.
(Ouyang et al., 2025) Code2MCP: automated transformation of code repositories into MCP services.
(Pan et al., 2025) Experiences with MCP servers for science and HPC.
(Anthropic, 2025) Claude Code overview.
(GTEx Consortium, 2020) GTEx portal.

Turning Papers Into Agents: Inside Paper2Agent’s MCP-Native Workflow
Paper2Agent: Reimagining Research Papers As Interactive and Reliable AI Agents