Cancer cells often rely on proteins that don't behave like typical enzymes to survive and spread. One such protein is CHI3L1 (also called YKL-40), which cancer cells secrete to communicate with their surroundings. Though CHI3L1 can't break down molecules like most enzymes, it still drives dangerous processes in cancer and inflammation. When CHI3L1 levels are high in patients, it typically signals a worse prognosis across multiple cancer types.
In brain cancer (glioblastoma or GBM) for example, CHI3L1 helps tumors become more aggressive and resistant to treatment. It activates several cellular pathways including JAK/STAT and PI3K/AKT, which promote tumor growth and help cancer cells evade the immune system.
Despite its clear importance in cancer, finding drugs that block CHI3L1 has been challenging. Most current CHI3L1 blockers are antibodies rather than small-molecule drugs, and few have been rigorously tested for how tightly they bind.
Now, researchers at Weill Cornell Medicine have developed a new approach to find CHI3L1-blocking compounds. They used a technique called surface plasmon resonance (SPR) to screen 704 potential drug molecules in real-time. This method identified two promising compounds that bind to CHI3L1 and demonstrated their effectiveness in a lab model that mimics brain tumors (Zhang et al., 2025).
Key Takeaways
- SPR-HTS of 704 compounds yielded two confirmed CHI3L1 binders (compounds 1-4 and 1-7) with Kd ≈ 10.4 μM and 7.4 μM, respectively (single-cycle kinetics).
- Both compounds disrupted the CHI3L1–galectin-3 interaction in AlphaLISA with IC50 ≈ 23.5 μM (1-4) and 15.4 μM (1-7).
- Docking and molecular dynamics placed the ligands in the CHI3L1 pocket with stabilizing hydrogen bonds; 1-7 showed the most stable binding trajectory.
- In a 3D GBM spheroid model that includes tumor cells, endothelium, and macrophages, compound 1-7 reduced viability and suppressed phospho-STAT3 more strongly than 1-4 and hygromycin B (HB), a reference CHI3L1–STAT3 disruptor.
- SPR provided kinetic heuristics (occupancy, early association/dissociation) that helped filter out non-specific binders at the primary screen stage.
Overview
CHI3L1 belongs to a family of proteins called glycoside hydrolases, but unlike its relatives, it can't break down molecules due to mutations in its active site. Instead, it works like a molecular messenger, binding to cellular receptors such as IL-13Rα2, galectin-3, and TMEM219. When CHI3L1 latches onto these receptors, it switches on multiple signaling pathways (ERK, AKT, NF-κB, WNT/β-catenin, and STAT3) that help cancer cells survive and multiply.
High levels of CHI3L1 in blood and tumor tissue have been documented across many cancer types and consistently predict poorer patient outcomes. In brain tumors specifically, CHI3L1 marks the most aggressive subtype and correlates with worse survival (Iwamoto & Hormigo, 2014; Wang et al., 2018).
Previous studies using antibodies to block CHI3L1 have shown promise, they effectively reduce tumor growth and spread in animal models, partly by preventing immune cells from switching to a tumor-friendly state and by dampening STAT3 activity.
However, developing small-molecule drugs (which are typically easier to manufacture and deliver than antibodies) has lagged behind. While some compounds like K284-6111 have shown activity in laboratory and animal studies, most lack the rigorous binding measurements needed for drug development. The current study fills this gap by using SPR, an optical technique that monitors how molecules bind to targets in real-time.
Why This Approach Matters
Finding drugs for targets like CHI3L1 is particularly challenging because traditional drug screening methods rely on measuring enzyme activity - but CHI3L1 doesn't function as an enzyme. SPR solves this problem by directly measuring whether potential drugs physically stick to their target protein. Unlike fluorescence-based methods, SPR isn't fooled by compounds that glow or interfere with light-based measurements.
More importantly, SPR reveals not just whether a compound binds, but how quickly it attaches and detaches from its target. This kinetic information helps researchers distinguish between compounds that bind specifically versus those that stick to everything, a common problem in drug screening. By establishing this SPR-based approach for CHI3L1, the researchers have created a path for systematic drug development against a validated cancer target.
The brain tumor spheroid experiments provide crucial evidence that CHI3L1 binding can translate into real biological effects. When the compounds successfully reduced tumor cell survival and blocked STAT3 signaling in a 3D model that includes multiple cell types, it demonstrated therapeutic potential beyond simple binding.
Results and Key Findings
The researchers started by testing 704 diverse compounds using a Biacore 8K SPR system. They attached CHI3L1 protein to a sensor chip and flowed each potential drug over it at a concentration of 100 μM.
The system measured four key parameters: how much of the target protein got occupied, how quickly compounds attached initially, and how rapidly they fell off during two different wash periods. Most compounds were eliminated because they either didn't bind strongly enough or stuck to everything non-specifically. Only two compounds, designated 1-4 and 1-7, passed the screening criteria.
Next, the team performed detailed binding measurements on the two winners. Both compounds showed rapid association with CHI3L1 followed by measurable dissociation, allowing calculation of their binding strengths (Kd values). Compound 1-4 bound with a Kd of approximately 10.4 μM, while compound 1-7 was slightly stronger at 7.4 μM. These measurements provide the quantitative foundation that's been missing for most reported CHI3L1 inhibitors.
To confirm the compounds actually interfere with CHI3L1's biological function, the researchers tested whether they could block CHI3L1 from binding to galectin-3, one of its natural partners. Using an AlphaLISA assay, they found that both compounds disrupted this interaction with IC50 values of 23.5 μM (1-4) and 15.4 μM (1-7). The fact that compound 1-7 performed better in both binding strength and biological activity suggests the measurements are capturing real drug-target interactions.
Computer modeling provided further insights into how the compounds might interact with CHI3L1 at the molecular level. Docking simulations placed both molecules in CHI3L1's binding pocket, with compound 1-4 forming a hydrogen bond with amino acid Asp207 and compound 1-7 engaging Trp99. Molecular dynamics simulations over time suggested that 1-7 maintained more stable contacts, consistent with its superior experimental performance.
The most compelling evidence came from testing the compounds in 3D brain tumor spheroids, lab-grown tumor models that include not just cancer cells, but also blood vessel cells and immune cells like those found in real tumors.
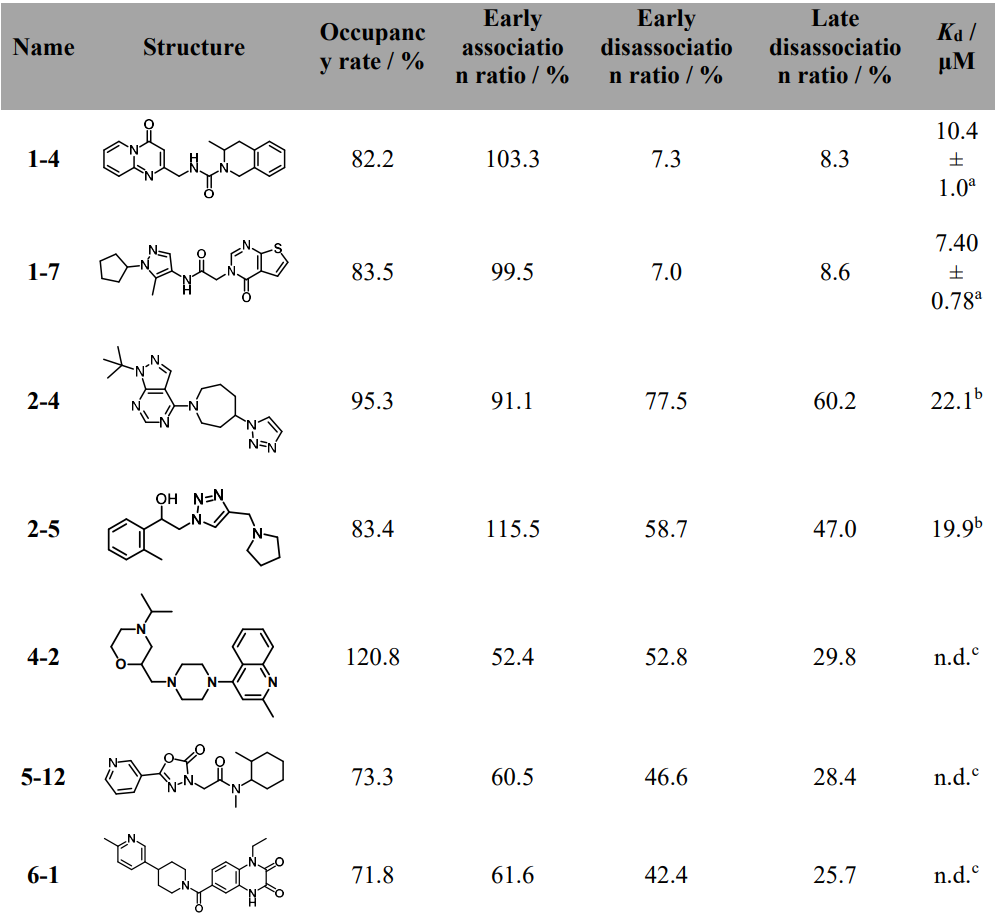
Table 1. Chemical structures, occupancy rate, early association ratio, early dissociation ratio, late dissociation ratio, and Kd values of 7 hits identified in primary screening.
Compound 1-7 reduced spheroid survival in a dose-dependent manner and significantly decreased levels of phosphorylated STAT3, a key protein that promotes tumor growth and immune suppression. Importantly, it outperformed both compound 1-4 and hygromycin B, a previously reported CHI3L1-STAT3 inhibitor, demonstrating clear therapeutic potential.
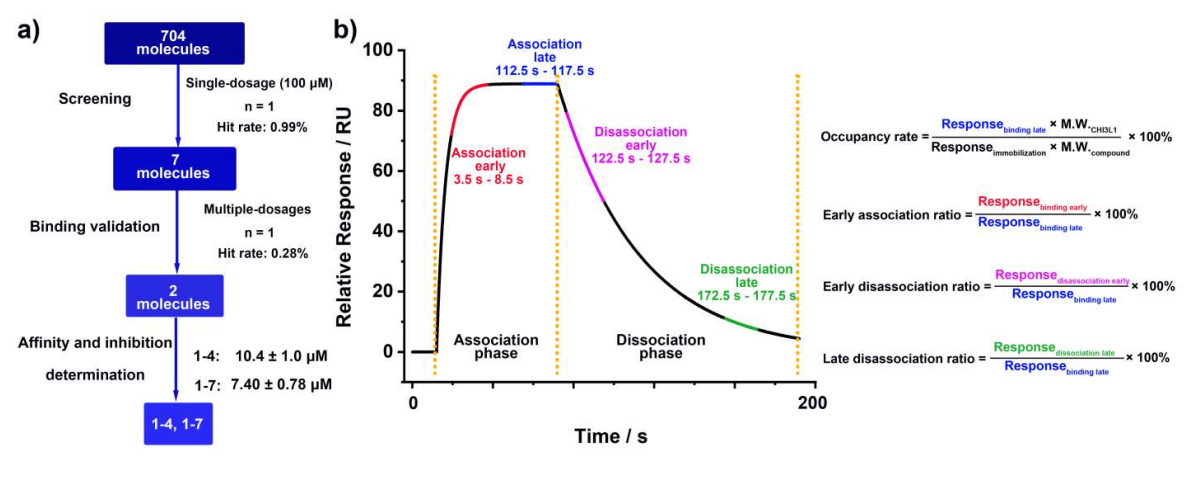
Figure 1. SPR-based HTS workflow and sensorgram analysis. a) Schematic illustration of the overall SPR-HTS pipeline used to identify CHI3L1 binders. A focused library of 704 molecules was screened at a single concentration (100 μM), yielding seven preliminary hits. Subsequent multi-concentration validation reduced this to two confirmed binders, compounds 1-4 and 1-7. b) Representative sensorgram illustrating association and dissociation phases, with time windows for early and late association/dissociation highlighted in red, blue, purple, and green, respectively. Quantitative parameters used for hit evaluation are shown on the right: occupancy rate, early association ratio, early dissociation ratio, and late dissociation ratio.
Figure 1 outlines the screening funnel on a Biacore 8K: 704 Discovery Diversity Set compounds injected at 100 μM over immobilized CHI3L1, with four metrics calculated from the sensorgrams - occupancy rate, early association ratio, early dissociation ratio, and late dissociation ratio. Most preliminary actives were eliminated due to rapid off-rates or persistent non-specific binding; two chemotypes (1-4, 1-7) advanced.
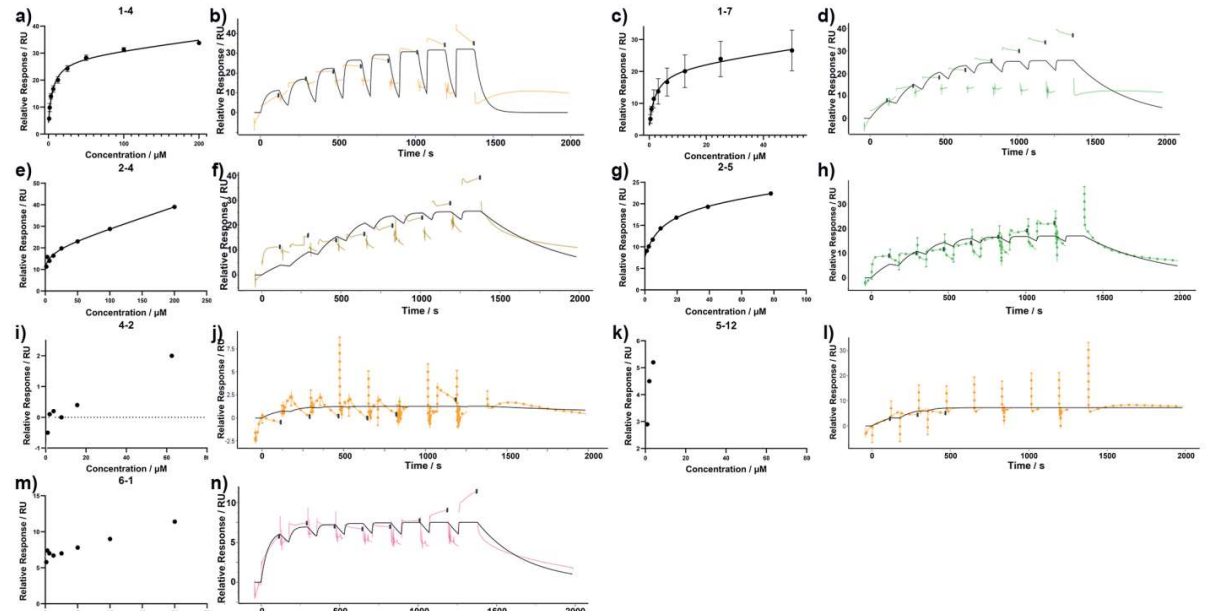
Figure 2. CHI3L1 binding affinity measurement using SPR. SPR-derived plots of relative response versus concentration (left panels) and sensorgrams (right panels) for compounds 1-4 (a, b), 1-7 (c, d), 2-4 (e, f), 2-5 (g, h), 4-2 (i, j), 5-12 (k, l), and 6-1 (m, n). Experiments for 1-4 and 1-7 were performed in triplicate, with results given as mean ± SD, whereas 2-4, 2-5, 4-2, 5-12, and 6-1 were tested once and reported as exact values.
In secondary single-cycle kinetics, compounds 1-4 and 1-7 showed rapid association to CHI3L1 and measurable dissociation phases suitable for Kd estimation, yielding ~10.4 μM and ~7.4 μM, respectively (Figure 2). These values provide a quantitative baseline that has been missing for many prior CHI3L1 ligands and enable rank-ordering for SAR.
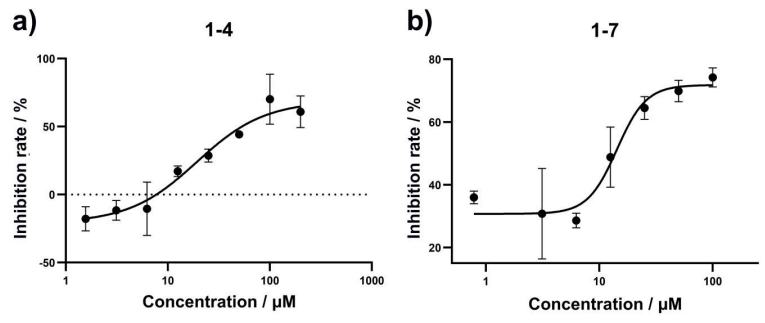
Figure 3. AlphaLISA-based inhibition assay. Inhibition curve of 1-4 (a) and 1-7 (b) against CHI3L1/galectin-3 interaction using the AlphaLISA inhibition assay. The inhibition rates were measured in triplicate with results given as the mean ± SD.
Orthogonal biochemical validation used an AlphaLISA assay to measure the inhibition of CHI3L1-galectin-3 binding. Dose-response curves gave IC50 ~23.5 μM (1-4) and ~15.4 μM (1-7) (Figure 3). While IC50 and Kd are not directly comparable, the consistency in rank order supports a shared mechanism: the compounds likely interfere with a CHI3L1 site important for protein-protein interaction.
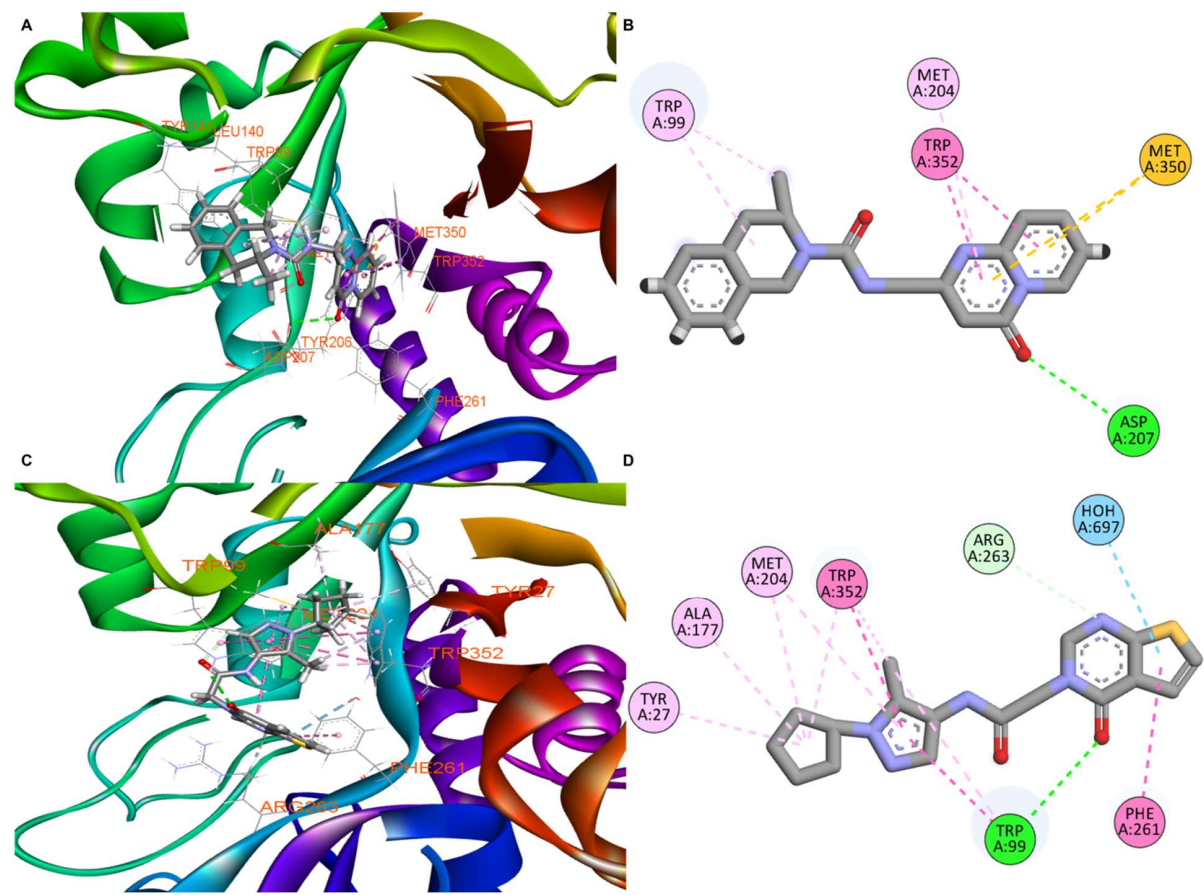
Figure 4. Molecular docking results of compounds 1-4 and 1-7 with CHI3L1. (a) 3D binding pose of compound 1-4 within the CHI3L1 binding pocket (PDB ID: 8R4X), showing key protein-ligand interactions. (b) 2D interaction diagram for compound 1-4, illustrating hydrogen bonds (green dashed lines), π-π stacking interactions (pink dashed lines), and hydrophobic contacts with labeled residues. (c) 3D binding pose of compound 1-7 within the CHI3L1 binding pocket, displaying the alternative binding orientation and interaction network. (d) 2D interaction diagram for compound 1-7, showing the distinct pattern of intermolecular interactions including hydrogen bonds, aromatic interactions, and hydrophobic contacts.
Docking and molecular dynamics placed both ligands in the CHI3L1 binding pocket. The 1-4 pose formed a hydrogen bond with Asp207 and hydrophobic contacts; 1-7 engaged Trp99 and a hydrophobic cleft (Figure 4).
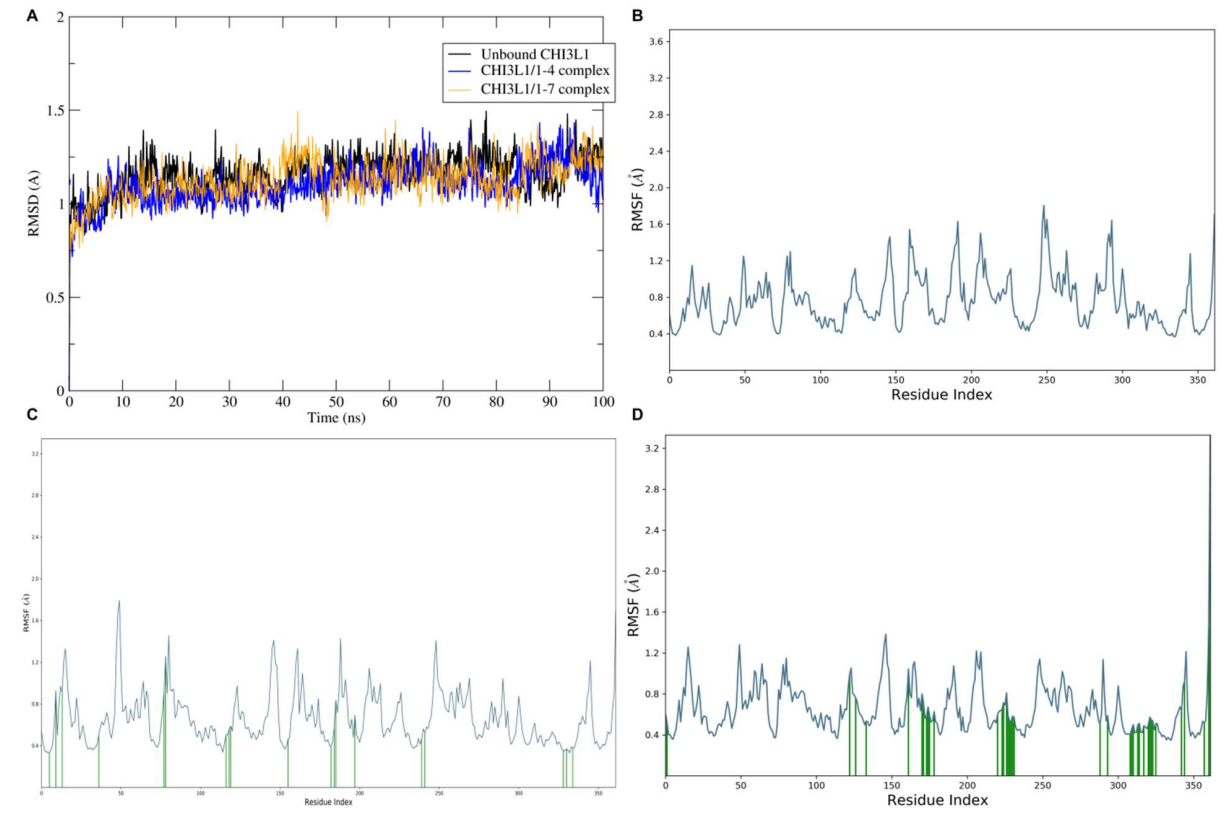
Figure 5. Molecular dynamics simulation analysis of CHI3L1 in different binding states. (a) Root mean square deviation (RMSD) over time showing structural stability of unbound CHI3L1 (black), CHI3L1/1-4 complex (blue), and CHI3L1/1-7 complex (orange) during 100 ns simulations. (b) Root mean square fluctuation (RMSF) per residue for unbound CHI3L1. (c) RMSF per residue for CHI3L1/1-4 complex. (d) RMSF per residue for CHI3L1/1-7 complex.
MD trajectories showed stable RMSD plateaus and persistent contacts for 1-7, suggesting a more conformationally locked pose (Figure 5). These in silico results are hypothesis-generating and align with the kinetic rank order.
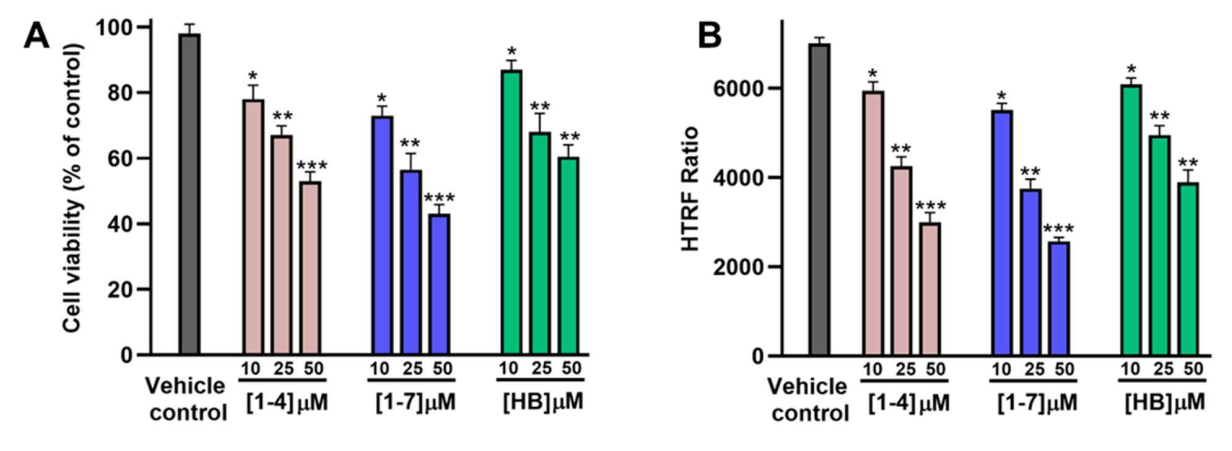
Figure 6. Evaluation of the therapeutic potential of compounds 1-4 and 1-7 in GBM spheroids. (A) Cell viability of GBM spheroids (as % of untreated control) upon incubation with increasing concentrations of the tested compounds (1-4, 1-7, and HB) after 72 h incubation. (B) Reduction in phospho-STAT3 levels, as determined by HTRF phospho-STAT3 kit from Revvity (Cat# 62AT3PET), in GBM spheroids upon incubation with the tested compounds (1-4, 1-7, and HB) after 72 h incubation. * p < 0.05, ** p < 0.01, and *** p < 0.001 relative to vehicle control. Data are representative of three independent experiments.
To test pathway engagement in a more physiological setting, the authors used a multicellular GBM spheroid model composed of U-87 MG cells co-cultured with endothelial and macrophage populations. Compound 1-7 induced a concentration-dependent drop in spheroid viability and a marked reduction in phosphorylated STAT3 (HTRF), outperforming 1-4 and hygromycin B (HB), a reported CHI3L1-STAT3 disruptor (Figure 6).
The concordance between binding, PPI inhibition, and STAT3 suppression supports on-target activity. These data are consistent with prior literature linking CHI3L1 to STAT3-mediated immune suppression and mesenchymal programs in GBM (Iwamoto & Hormigo, 2014; Wang et al., 2018).
Study Limitations
While these results represent a significant step forward in CHI3L1 drug discovery, the study has several important limitations. The binding affinities measured (7-10 μM) fall in the moderate range and would likely need improvement for therapeutic applications. The biological validation was limited to one spheroid model and specific cell lines, so broader testing across diverse cancer types would strengthen confidence in the approach.
Additionally, while the compounds showed promising activity in laboratory conditions, their behavior in living organisms remains unknown - important factors like absorption, distribution, metabolism, and potential side effects need investigation. Future work should focus on optimizing these early compounds to achieve stronger binding, testing them in additional disease models, and evaluating their safety profiles.
Looking Forward
This research demonstrates that SPR-based screening can successfully identify promising drug candidates for CHI3L1, a challenging but important cancer target. The systematic approach - combining biophysical measurements, biological validation, and computational modeling - provides a robust foundation for future drug development efforts. The discovery of compounds that can measurably bind CHI3L1 and disrupt its cancer-promoting activities represents an important first step toward potential new treatments for aggressive brain tumors.
For researchers interested in cancer drug discovery, this work illustrates how modern screening technologies can tackle previously "undruggable" targets. The methods described here could be applied to other challenging proteins involved in cancer progression. If you're working in this field, the full preprint provides detailed protocols that could inform your own screening campaigns.
Definitions
CHI3L1/YKL-40: A secreted chitinase-like lectin implicated in cancer, neurodegeneration, and inflammation; non-enzymatic but pro-signaling.
SPR (Surface Plasmon Resonance): A label-free technique that measures real-time binding of molecules to immobilized targets by detecting refractive index changes at a sensor surface; outputs association/dissociation curves and kinetic parameters.
AlphaLISA: A bead‑based luminescent assay for detecting biomolecular interactions in homogeneous format; suitable for PPI inhibition measurements.
GBM Spheroid: A 3D co‑culture model that more closely mimics tumor architecture and microenvironment than 2D monolayers.
STAT3: A transcription factor activated downstream of cytokine and growth factor signaling; promotes immune suppression and survival in many cancers.
References
Zhang, L.; Hammouda, H. H. N.; Gabr, M. T. Discovery of Small Molecule CHI3L1 Inhibitors by SPR-Based High-Throughput Screening. bioRxiv, 2025. (Zhang et al., 2025).
Iwamoto, F. M.; Hormigo, A. Unveiling YKL-40, from Serum Marker to Target Therapy in Glioblastoma. Frontiers in Oncology, 2014. (Iwamoto & Hormigo, 2014).
Wang, Y. et al. Differential regulation of YKL-40/CHI3L1 by PTEN/PI3K and JAK2/STAT3 pathways in glioblastoma. Cancer Letters, 2018. (Wang et al., 2018).

SPR Finds Small-Molecule Binders For CHI3L1, A Hard Cancer Target
Discovery of Small Molecule CHI3L1 Inhibitors by SPR-Based High-Throughput Screening