Training capable biomedical language models hinges on having the right data: not just more tokens, but high-fidelity question-answer examples grounded in the structure of biomedical knowledge.
The paper by Xiao et al. proposes m-KAILIN, a knowledge-driven, agentic framework that distills AI-ready datasets directly from scientific literature. Instead of manual curation or single-agent synthesis, the authors orchestrate specialized agents that generate questions, retrieve context, evaluate alignment using the Medical Subject Headings (MeSH) hierarchy, and produce answers, yielding question-context-answer triples tuned for downstream QA training.
The authors claim that models trained on m-KAILIN distilled corpora achieve notable gains on PubMedQA and, at large scale, Llama 3 70B surpasses GPT-4 with MedPrompt and Med-PaLM-2 in the reported setup.
The method emphasizes ontology guidance and multi-agent diversity over sheer parameter count, aligning with recent retrieval-augmented and preference-optimized trends in domain modeling (Karpukhin et al., 2020), (Lewis et al., 2020), (Dubey et al., 2024), (Labrak et al., 2024), (Achiam et al., 2023), (Singhal et al., 2023).
Key Takeaways
- Multi-agent pipeline with distinct Question Generation Agents, domain retrieval, and an ontology-guided Evaluation Agent produces high-precision QA data aligned to MeSH.
- Knowledge-guided cold-start labels train an LLM evaluator, replacing human preferences with MeSH-based similarity to select better question-context pairs.
- On PubMedQA, m-KAILIN improves both small and large models; Llama 3 70B trained on m-KAILIN data edges past Med-PaLM-2 and GPT-4+MedPrompt in the paper’s experiments.
- Ablations show three levers matter most: heterogeneous question generators for diversity, domain-adapted retrieval, and MeSH-aware evaluation.
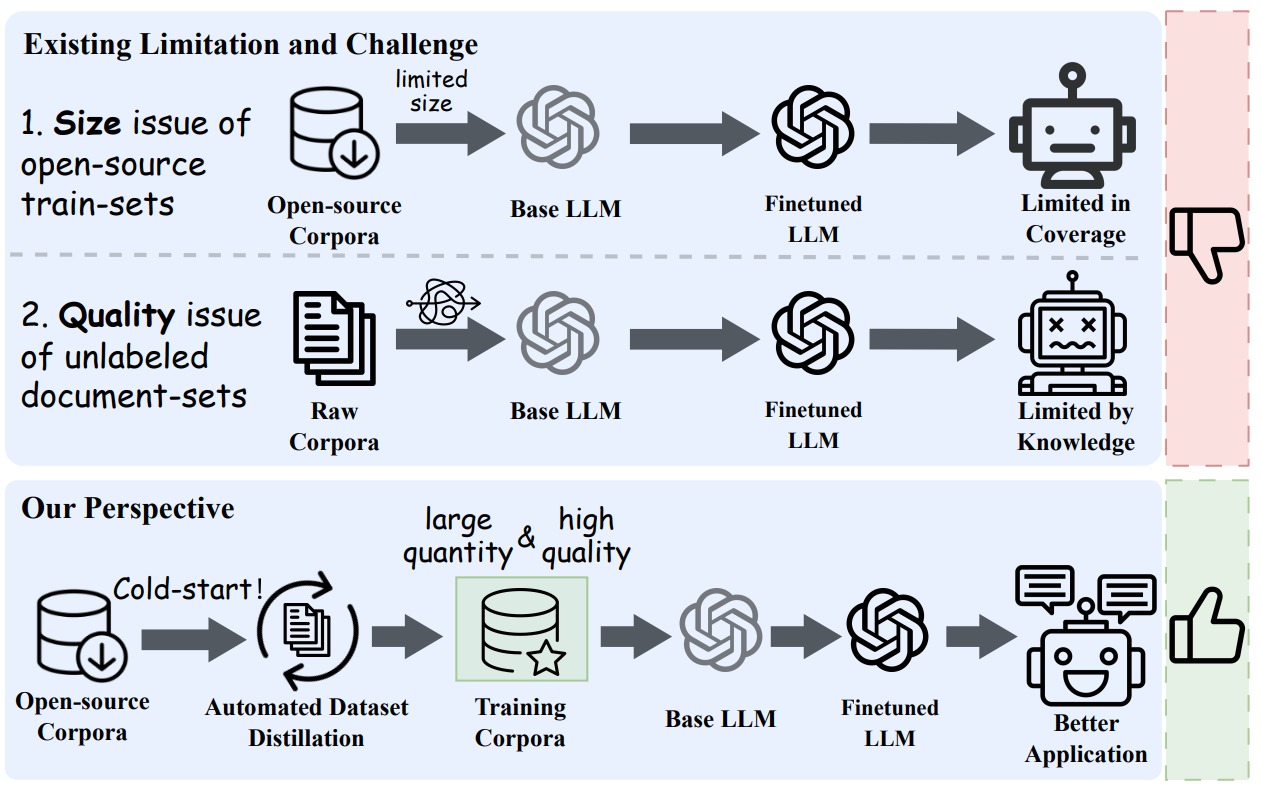
Fig. 1: Analyzing the limitations and challenges of the existing pipeline. The motivation of this study is to utilize the highquality but limited annotated corpus to generate large-scale training corpora from raw scientific documents. Credit: Xiao et al.
How the framework works
m-KAILIN decomposes corpus distillation into cooperating agents. Two Question Generation Agents are fine-tuned on biomedical QA to create candidate questions from raw papers: one uses a domain LLM such as BioMistral (Labrak et al., 2024), while the other uses a strong generalist such as Llama 3 (Dubey et al., 2024). This heterogeneity increases coverage and reduces redundancy in the question space.
For each question, a Context Retrieval Agent encodes the query and candidate passages with a biomedical encoder and selects the top-k contexts using dense retrieval, in the spirit of DPR and RAG (Karpukhin et al., 2020), (Lewis et al., 2020).
Candidate question-context pairs are then compared by a MeSH-guided Question Evaluation Agent that computes knowledge-hierarchy similarity using MeSH information content (Lipscomb, 2000). These rule-built labels train an LLM evaluator, removing the need for human preference annotation.
Once a preferred pair is chosen, an Answer Generation Agent produces an answer to build full question-context-answer triples. The authors then apply direct preference optimization to steer the generalist question generator toward preferred outputs (Rafailov et al., 2024).
The resulting data supports both continuous pre-training on question-context pairs and supervised fine-tuning on question-context-answer triples, matching downstream QA formats like PubMedQA (Jin et al., 2019).
Why it is important
Biomedical QA is brittle when models ignore domain structure. Ontologies like MeSH encode the hierarchy and semantics that clinicians and researchers rely on. Injecting that structure into evaluation helps the pipeline select questions with the right granularity and terminology, improving training signals without manual labeling. While retrieval anchors questions in evidence and reduces hallucinations when the model trains to produce answers consistent with source passages.
The broader implication is practical: high-quality, ontology-aligned corpora can allow smaller open models to compete with larger proprietary ones on targeted biomedical tasks. That has significant cost, access, and transparency benefits for labs that cannot train or deploy very large models. The framework may also generalize conceptually to other verticals with strong ontologies and taxonomies, such as chemistry or clinical coding systems.
Discussion of results
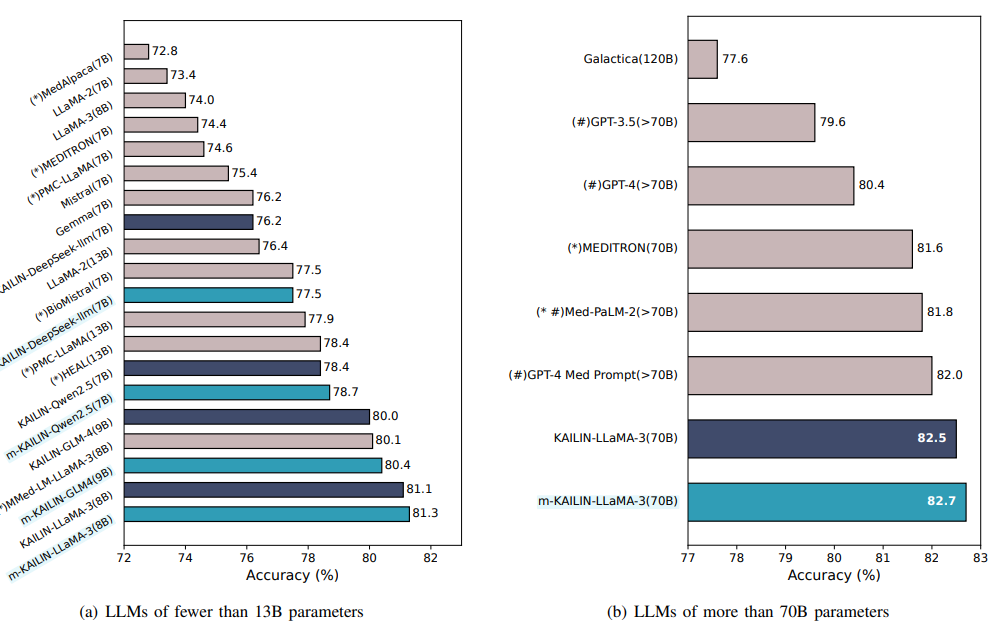
Figure 7 in the paper compares PubMedQA accuracy across two scale regimes. For models with fewer than 13B parameters, m-KAILIN consistently lifts accuracy, narrowing the gap with larger systems. In the 70B-plus group, Llama 3 70B trained on m-KAILIN data edges past GPT-4 with MedPrompt and Med-PaLM-2 in the authors’ setup, showing how data alignment can rival pure scale, (Achiam et al., 2023), (Singhal et al., 2023).
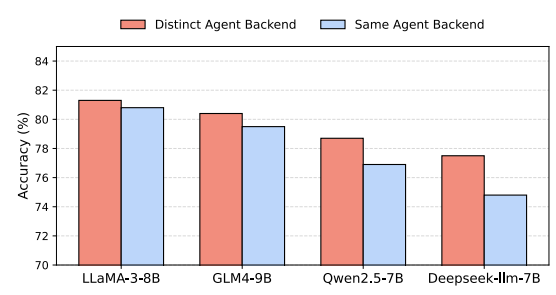
Figure 8 examines agent heterogeneity: pairing a domain specialist with a generalist outperforms using two generalists, indicating that diversity in question generation improves coverage and reduces mode collapse.
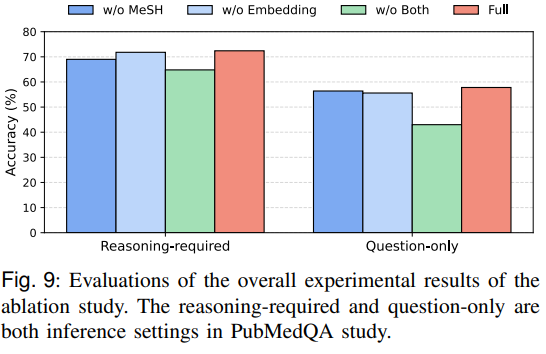
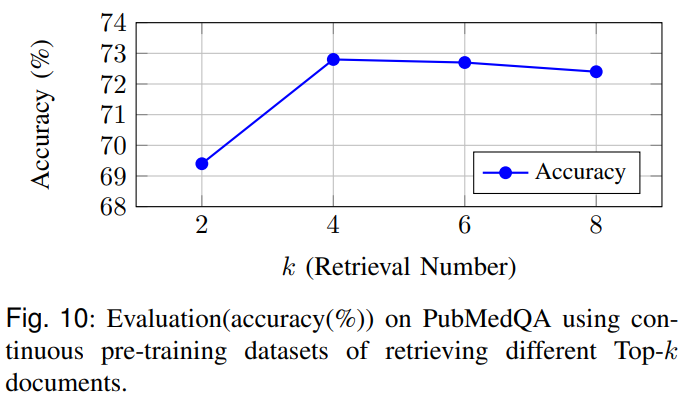
Figures 9 and 10 isolate retrieval and ontology guidance: removing MeSH guidance or replacing domain embeddings with generic ones reduces accuracy; an ablation on the retrieval top-k finds k=4 balances context richness with noise in continuous pre-training.
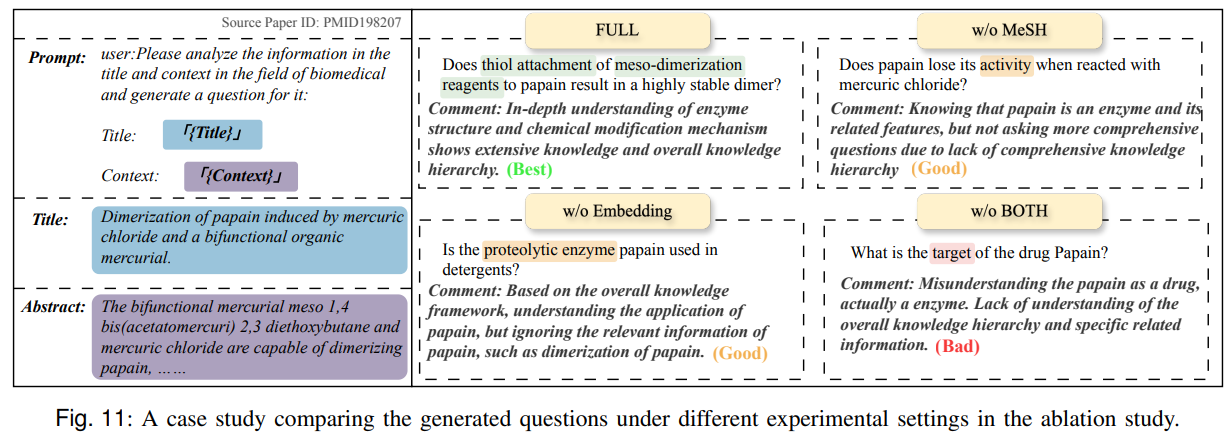
Figure 11’s case analysis shows that MeSH-aware evaluation selects questions and contexts that more accurately capture biochemical roles and relations, while generic embeddings miss fine-grained cues.
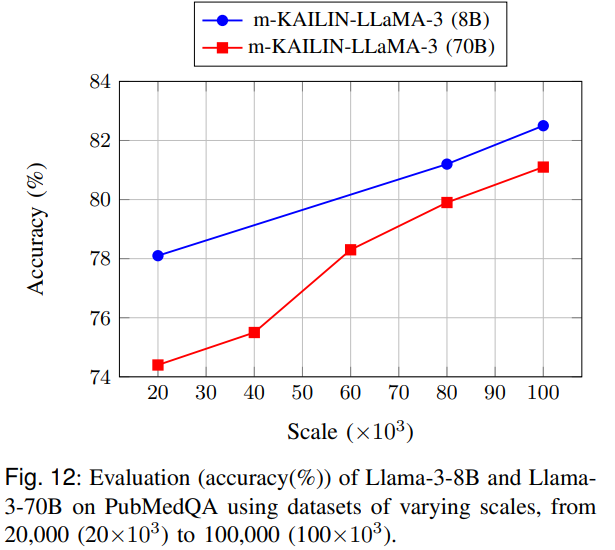
Figure 12 studies scaling laws: both 8B and 70B models improve as the distilled dataset grows from 20k to 100k examples, with diminishing returns for the larger model. That suggests smaller models may benefit disproportionately from better-aligned data when task complexity is bounded.
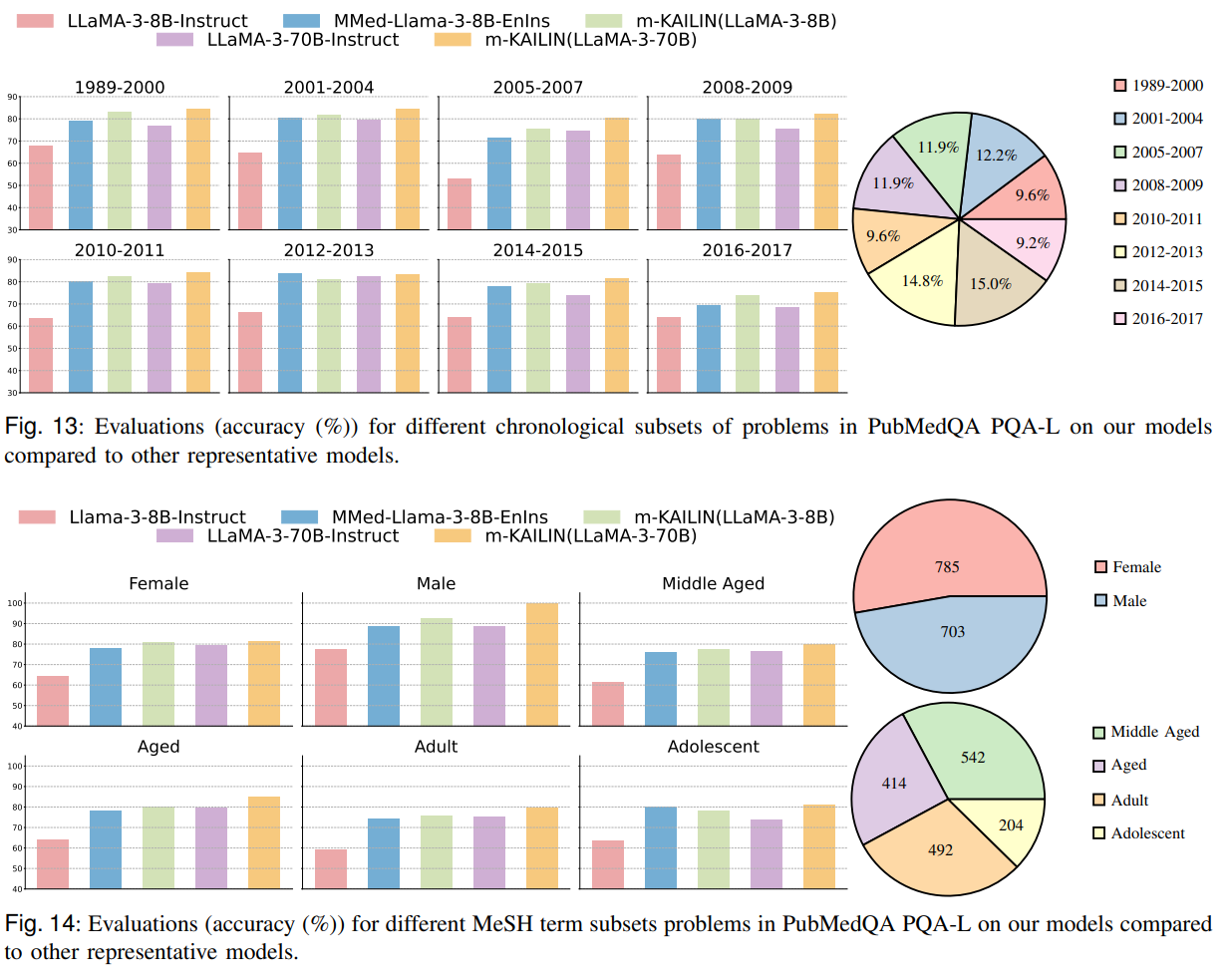
Figures 13 and 14 slice robustness by publication time and MeSH subdisciplines; m-KAILIN-trained models show steadier performance across eras and topics, consistent with the idea that ontology-aware selection improves generalization across shifting biomedical vocabularies and demographics.
Conclusion
m-KAILIN is a strong blueprint for scientific corpus distillation in biomedicine: combine heterogeneous question generators, retrieval, and MeSH-guided evaluation, then optimize with preference training to produce AI-ready QA corpora.
The reported gains on PubMedQA point to a clear lesson: structure and selection can substitute for scale, especially when domain ontologies are available. For teams building biomedical QA systems, starting with ontology-guided evaluation and domain retrieval is an actionable path to better data and better models.
Further work should probe external reproducibility, evaluate on additional biomedical QA suites, and test portability to other ontology-rich domains. The authors share pointers to data and prompts in the paper Xiao et al., 2025.

m-KAILIN Turns Biomedical Literature into AI-ready QA Data
m-KAILIN: Knowledge-Driven Agentic Scientific Corpus Distillation Framework for Biomedical Large Language Models Training