Medical image semantic segmentation is a fundamental process in modern healthcare, crucial for applications such as diagnosing diseases, tracking disease progression, planning treatments, and assisting surgeries.
GenSeg, a generative AI framework, is tackling one of the biggest obstacles in medical deep learning: the scarcity of expert-annotated data for image segmentation. GenSeg creates synthetic, high-fidelity image-mask pairs, enabling segmentation models to thrive even in environments where annotated images are rare allowing accurate medical image segmentation with only a handful of labeled examples.
Key Takeaways:
- Overcoming Data Scarcity: Deep learning models for medical image segmentation typically require vast, meticulously annotated datasets, which are costly and time-consuming to obtain, leading to "ultra low-data regimes" where models struggle to generalize.
- Generative AI Solution: GenSeg introduces a novel generative deep learning framework that creates high-fidelity synthetic medical images paired with their corresponding segmentation masks, acting as auxiliary training data.
- End-to-End Multi-Level Optimization (MLO): Unlike traditional generative models that separate data generation from model training, GenSeg uses a unique MLO process that allows the performance of the segmentation model to directly guide and optimize the data generation process, ensuring the synthetic data is highly effective for improving segmentation outcomes.
- Superior Performance: GenSeg significantly boosts segmentation performance by 10–20% (absolute gains) in scenarios with limited real data, performing well in both "in-domain" (training and testing data from the same distribution) and "out-of-domain" (OOD, training and testing data from different distributions) settings.
- Enhanced Data Efficiency: The framework requires 8–20 times less training data than existing approaches to achieve comparable or superior performance. This drastically reduces the burden and cost associated with medical image annotation.
- Robust Generalization: GenSeg improves segmentation models' ability to generalize to new, unseen data, even from different domains or imaging modalities, which is critical for real-world clinical applicability.
- Versatile and Model-Agnostic: GenSeg seamlessly integrates with diverse popular segmentation models like UNet, DeepLab, and Transformer-based SwinUnet, enhancing their capabilities. It has also been successfully extended to 3D medical image segmentation.
- Outperforms State-of-the-Art: The framework consistently outperforms widely used data augmentation methods, existing data generation tools (e.g., WGAN), semi-supervised learning techniques, and even the state-of-the-art medical image segmentation tool, nnUNet, especially in data-scarce environments.
Credit: Paper
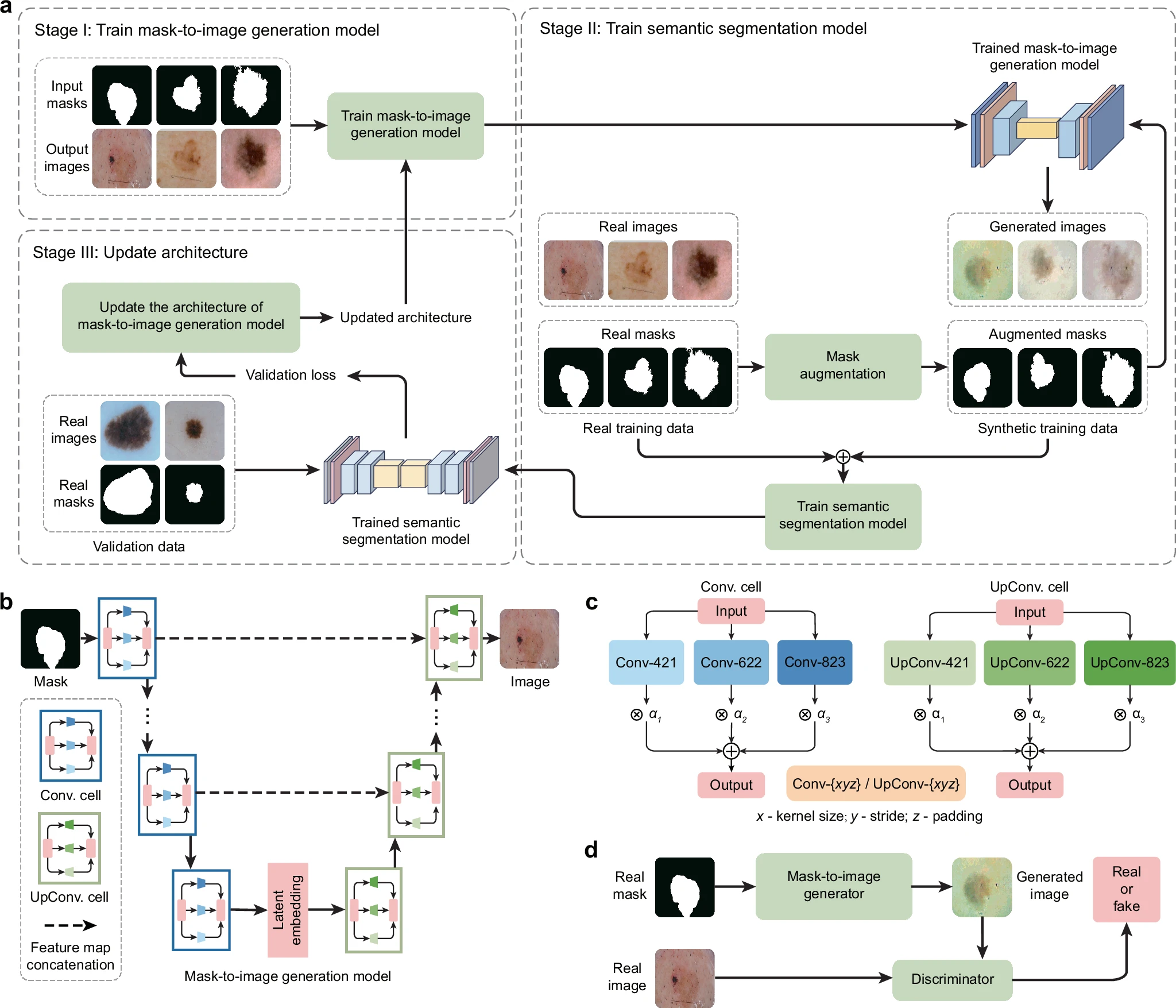
a Overview of the GenSeg framework. GenSeg consists of (1) a semantic segmentation model that predicts a segmentation mask from an input image, and (2) a mask-to-image generation model that synthesizes an image from a segmentation mask. The latter features a neural architecture that is both learnable in structure and parameterized by trainable weights. GenSeg operates through three end-to-end learning stages. In stage I, the network weights of the mask-to-image model are trained with real mask-image pairs, with its architecture tentatively fixed. Stage II involves using the trained mask-to-image model to synthesize training data. Real segmentation masks are augmented to create new masks, from which synthetic images are generated. These synthetic image-mask pairs are used alongside real data to train the segmentation model. In stage III, the trained segmentation model is evaluated on a real validation dataset, and the resulting validation loss—which reflects the performance of the mask-to-image model—is used to update this architecture. Following this update, the model re-enters Stage I for further training, and this cycle continues until convergence. b Searchable architecture of the mask-to-image generation model. It comprises an encoder and a decoder. The encoder processes an input mask into a latent representation using a series of searchable convolution (Conv.) cells. The decoder employs a stack of searchable up-convolution (UpConv.) cells to transform the latent representation into an output medical image. Each cell, as shown in (c) contains multiple candidate operations characterized by varying kernel sizes, strides, and padding options. Each operation is associated with a weight α denoting its importance. The architecture search process optimizes these weights, and only the most influential operations are retained in the final model. d The weight parameters of the mask-to-image generator are trained within a generative adversarial network (GAN) framework, in which a discriminator learns to distinguish real images from generated ones, while the generator is optimized to produce images that are indistinguishable from real images. All qualitative examples are sourced from publicly available datasets.
The Data Scarcity Challenge in Medical Imaging
Deep learning has transformed medical imaging, streamlining disease diagnosis, treatment planning, and surgical support. But these advances often stall due to the high cost and time required for assembling large, expertly labeled datasets. In these "ultra low-data regimes," models can overfit, leading to poor generalization on new cases.
GenSeg’s Integrated Generative Solution
Traditional approaches, like data augmentation and semi-supervised learning, either separate data creation from model training or require lots of unlabeled data, which is not always available due to privacy constraints. GenSeg addresses these gaps by unifying synthetic data generation and segmentation training in one end-to-end workflow.
- Reverse Generation Mechanism: GenSeg begins with a real annotated mask, augments it, and then uses a generative model to synthesize the matched medical image, maintaining high semantic accuracy.
- Adaptive Architecture: The generative model's structure is automatically learned, adapting to specific segmentation tasks and datasets.
- Multi-Level Optimization (MLO): Synced optimization of data generation and model performance ensures that both components improve together, creating a robust feedback loop.
Efficiency and Performance Gains
Across diverse segmentation tasks, GenSeg consistently delivers 10–20% absolute performance improvements over standard models in low-data settings. It achieves similar or superior accuracy with 8–20 times fewer labeled examples, dramatically reducing annotation costs and development time.
- Superior Out-of-Domain Generalization: GenSeg-trained models retain strong performance on new datasets or imaging types, crucial for clinical adoption.
- Versatility: GenSeg integrates with popular segmentation networks, UNet, DeepLab, SwinUnet, and scales to 3D modalities like MRI and CT scans.
- Computational Efficiency: Training is fast (often under 2 GPU hours) and does not increase inference costs, enabling use in resource-constrained settings.
Beating the State of the Art
GenSeg outperforms leading data augmentation, generative models (e.g., WGAN), semi-supervised methods, and advanced segmentation architectures like nnUNet. Its holistic approach ensures that synthetic data is both realistic and highly effective for training.
- For lung segmentation, GenSeg-UNet hit a 94.5% Dice score with just 9 training examples, over 16% better than nnUNet.
- For skin lesion segmentation, it improved the Jaccard index from 0.41 to 0.65 on out-of-domain data, using far fewer labeled samples.
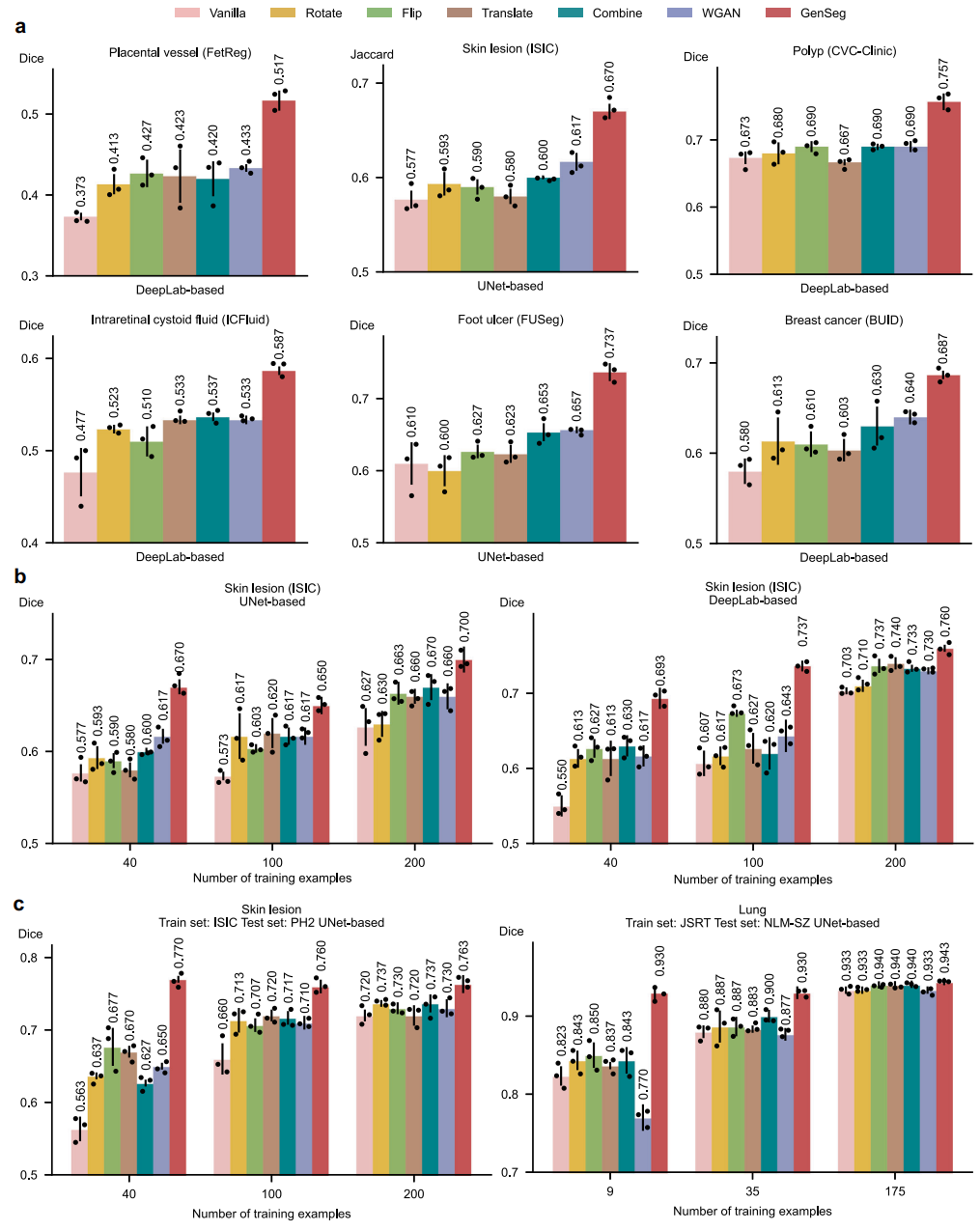
Fig. 5 | GenSeg significantly outperformed widely used data augmentation and generation methods. a GenSeg’s in-domain generalization performance compared to baseline methods, including Vanilla (without any data augmentations), Rotate, Flip, Translate, Combine, and WGAN, when used with UNet or DeepLab in segmenting placental vessels, skin lesions, polyps, intraretinal cystoid fluids, foot ulcers, and breast cancer using the FetReg, ISIC, CVC-Clinic, ICFluid, FUSeg, and BUID datasets. b GenSeg’s in-domain generalization performance compared to baseline methods using a varying number of training examples from the ISIC dataset for segmenting skin lesions, with UNet and DeepLab as the backbone segmentation models. c GenSeg’s out-of-domain generalization performance compared to baseline methods across varying numbers of training examples in segmenting lungs (using examples from JSRT for training, and NLM-SZ and NLMMC for testing) and skin lesions (using examples from ISIC for training, and DermIS and PH2 for testing), with UNet and DeepLab as the backbone segmentation models. In all panels, bar heights represent the mean, and error bars indicate the standard deviation across three independent runs with different random seeds. Results from individual runs are shown as dot points. Source data are provided as a Source Data file.
Technical Innovations Driving GenSeg
- Mask-First Generation: Starting from the mask guarantees that generated images align closely with clinical labels.
- Learnable Generator Architectures: Automated architecture search tailors the generator to each dataset’s unique characteristics.
- Optimized Augmentation: Strategic combinations of image transformations maximize out-of-domain model robustness.
Limitations and Next Steps
GenSeg’s performance depends on the quality and diversity of the initial annotated images; any biases may be inherited by the synthetic data. While GenSeg generalizes well, its performance could decline with radically different imaging modalities.
Future work includes improving synthetic data fidelity, learning from even fewer samples, and expanding to tasks like anomaly detection or multimodal analysis. Incorporating clinical expert feedback will also ensure clinical relevance and minimize artifacts.
The Takeaway: Transforming Medical AI Accessibility
GenSeg represents a significant leap for medical image segmentation in data-scarce environments. By tightly integrating data generation and model training, it delivers robust, scalable, and cost-effective segmentation solutions. As AI continues to reshape healthcare, GenSeg’s innovations are poised to drive wider access and higher-quality medical image analysis tools.

GenSeg For Medical Image Segmentation in Low-Data Environments
Generative AI enables medical image segmentation in ultra low-data regimes