In a groundbreaking fusion of paleobiology and artificial intelligence, researchers from the University of Pennsylvania have pioneered a revolutionary approach to combat the escalating global antibiotic resistance crisis. Their study, published in Nature Biomedical Engineering, introduces the concept of "molecular de-extinction" - the resurrection of antimicrobial molecules from the proteomes of extinct organisms using deep learning algorithms.
Led by Cesar de la Fuente-Nunez and his team, this research represents a paradigm shift in drug discovery, tapping into an evolutionary reservoir of antimicrobial compounds that vanished with their hosts but remain encoded in their genetic blueprints.
The urgency of this work cannot be overstated. According to the World Health Organization, antimicrobial resistance was directly responsible for 1.27 million global deaths in 2019 and contributed to 4.95 million deaths.
Without effective intervention, projections suggest antimicrobial-resistant infections could cause 10 million annual fatalities by 2050. The researchers addressed this crisis by developing APEX (Antibiotic Peptide de-Extinction), a sophisticated deep learning framework that systematically mined the proteomes of 208 extinct species, analyzing over 10 million peptide sequences to identify those with potent antimicrobial properties.
Key Takeaways
- APEX algorithm successfully identified 37,176 peptides with predicted broad-spectrum antimicrobial activity from extinct organisms, with 11,035 unique to extinct species
- 69 synthesized peptides showed a 59% hit rate for antimicrobial activity, significantly outperforming previous scoring methods (24%)
- Lead compounds from woolly mammoths (mammuthusin-2), straight-tusked elephants (elephasin-2), giant sloths (mylodonin-2), and other extinct species demonstrated therapeutic efficacy comparable to polymyxin B in mouse infection models
- Unlike conventional antimicrobial peptides that target outer membranes, these ancient peptides primarily kill bacteria through cytoplasmic membrane depolarization
- Archaic encrypted peptides (AEPs) displayed distinct physicochemical properties, including higher uncharged polar residue content and reduced amphiphilicity compared to modern antimicrobials
- Multiple peptide combinations showed synergistic effects, with some reducing minimum inhibitory concentrations by up to 64-fold
Decoding Ancient Immunity Through Deep Learning
The APEX algorithm represents a significant advancement in computational antimicrobial discovery. Traditional approaches to identifying antimicrobial peptides have relied heavily on physicochemical scoring functions that prioritize net positive charge and amphiphilicity ( having both a polar (hydrophilic) and a nonpolar (hydrophobic) part).
However, APEX employs a multitask deep learning architecture that combines recurrent neural networks with attention mechanisms to extract complex patterns from peptide sequences. The model was trained on a comprehensive dataset comprising 988 in-house peptides with 14,738 antimicrobial activity measurements across 34 bacterial strains, augmented with 5,093 publicly available antimicrobial peptides and 5,500 inactive controls from the Database of Antimicrobial Activity and Structure of Peptides (DBAASP).
This sophisticated approach enabled APEX to identify antimicrobial candidates that conventional methods would likely overlook. The algorithm's ensemble learning strategy, which averages predictions from eight top-performing models with different neural network architectures, achieved performance metrics: R² values of 0.546, Pearson correlations of 0.728, and Spearman correlations of 0.607 when evaluated on independent test sets. These results substantially outperformed baseline machine learning methods including random forests, gradient boosting, and support vector regression.
The deep learning framework also incorporated phylogenetic constraints, encouraging the model to give similar predictions for closely related bacterial species. This biological insight proved valuable, as six of the eight selected models utilized this constraint during training.
The multitask learning approach, which simultaneously predicted species-specific antimicrobial activities and performed binary classification of antimicrobial versus non-antimicrobial peptides, served as an effective data augmentation strategy that improved overall prediction performance.
Why Ancient Molecules Matter for Modern Medicine
The concept of molecular de-extinction addresses a critical limitation in current drug discovery: the finite exploration of molecular diversity. While modern drug development has largely focused on synthetic compounds and molecules from extant organisms, this approach leaves vast swaths of evolutionary chemical space unexplored.
Extinct organisms represent billions of years of evolutionary innovation, during which natural selection optimized molecular solutions for survival challenges including microbial threats. By accessing this "extinctome," researchers can potentially discover entirely new classes of antimicrobial compounds with novel mechanisms of action.
The study's findings reveal that antimicrobial peptides from extinct organisms occupy distinct regions of chemical space compared to known antimicrobials. Using uniform manifold approximation and projection (UMAP) analysis, the researchers demonstrated that APEX-identified sequences formed multiple distinct clusters not covered by existing antimicrobial databases. This suggests that molecular de-extinction can genuinely expand our arsenal of therapeutic compounds rather than simply rediscovering known antimicrobials.
The evolutionary perspective also provides insights into the origins of innate immunity. The widespread presence of antimicrobial peptides across extinct species suggests that these molecules played crucial roles in host defense throughout evolutionary history.
Understanding how ancient organisms defended against microbial threats could inform the development of next-generation immunotherapies and preventive strategies. Moreover, the success of these ancient antimicrobials validates the hypothesis that natural selection has been conducting drug discovery experiments for millions of years, with results now accessible through computational archaeology.
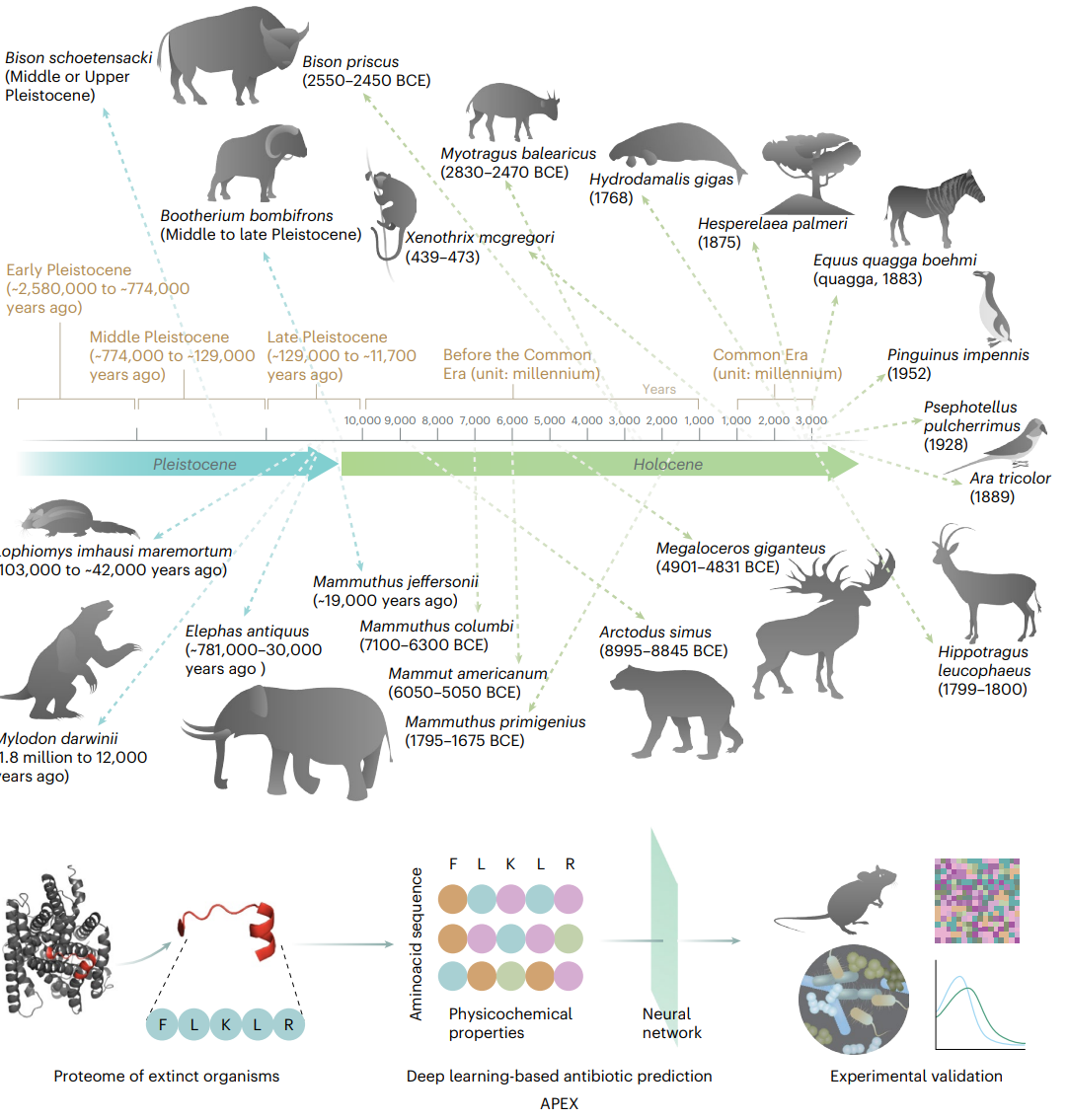
Fig. 1 | Molecular de-extinction of antibiotics from ancient proteomes using deep learning. All available proteomes of extinct organisms were mined by APEX deep learning algorithm. Amino acid sequences ranging from 8 to 50 amino acid residues within proteins from extinct organisms were inputted into multitask deep learning models that trained on both public and in-house peptide data to evaluate the potential antimicrobial activity. The highest ranked peptides based on predicted antimicrobial activities were then selected and thoroughly characterized against clinically relevant pathogens both in vitro and in animal models. The mechanism of action, physicochemical features and synergistic interactions of these peptides were also assayed. The dates report the approximate extinction date or period for the organisms studied. The protein and peptide structures shown in the figure were created with PyMOL Molecular Graphics System, version 2.1 Schrödinger, LLC. Credit: wan et al.
Experimental Validation Reveals Unique Properties
The transition from computational prediction to experimental validation demonstrated the practical potential of molecular de-extinction. Of the 69 peptides synthesized from extinct organisms, 41 showed notable antimicrobial activity against at least one of 11 clinically relevant bacterial pathogens, including dangerous multidrug-resistant strains such as vancomycin-resistant Enterococcus faecium and methicillin-resistant Staphylococcus aureus. The 59% hit rate substantially exceeded that of previous scoring methods applied to the same extinct sources, underscoring the superiority of the deep learning approach.
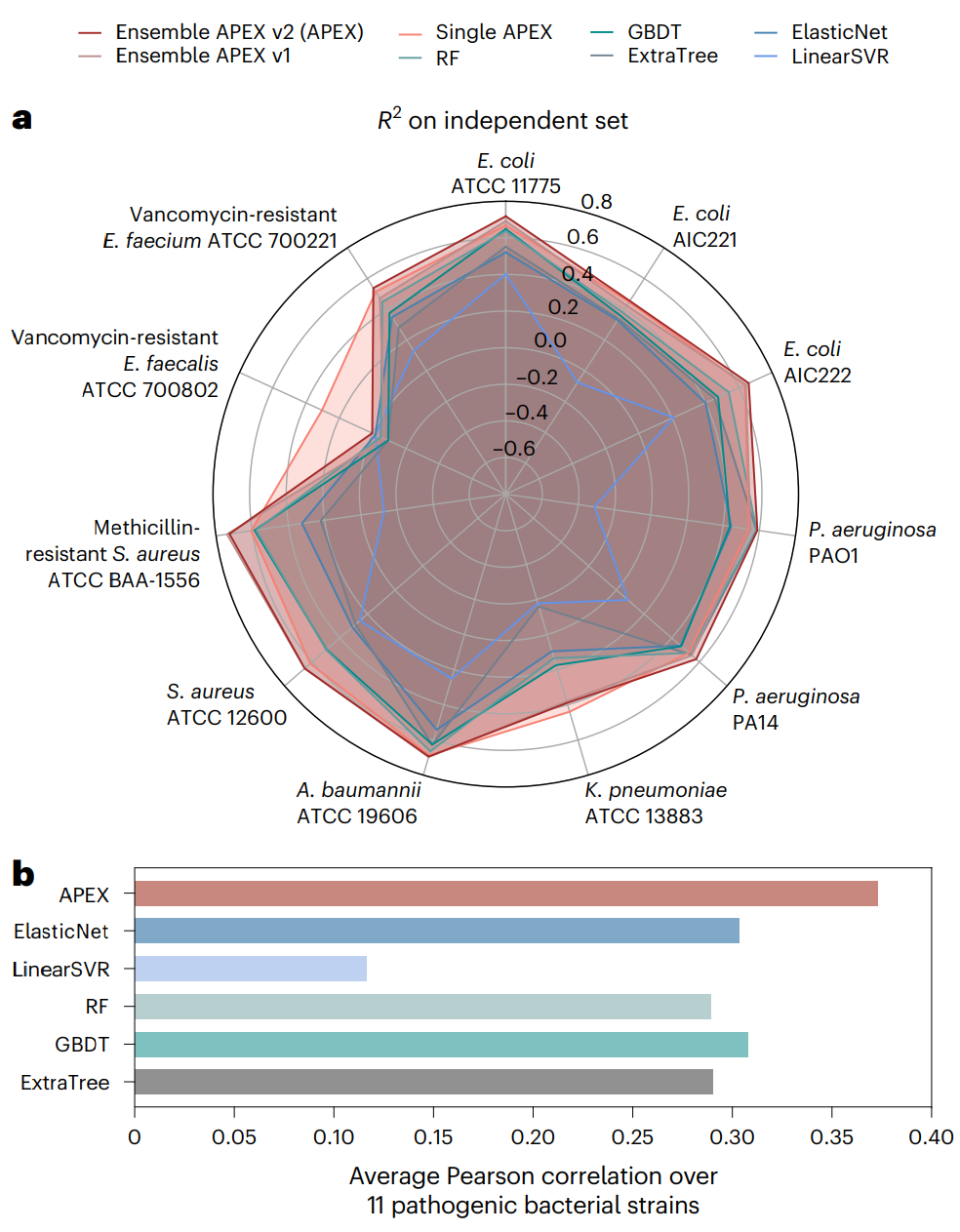
Fig. 2 | APEX prediction performance and comparison with other models. a, Radar chart showing R2 correlation in terms of species-specific antimicrobial activity prediction on an independent dataset (a held-out subset from our inhouse peptide dataset) for various ML models. The radius reflects the R2 value for each of the models. APEX variants outperformed the baseline ML methods for most of the pathogens analysed. RF, random forest; GBDT, gradient boosting decision tree; ExtraTree, extra-tree regressor; ElasticNet, elastic net; LinearSVR, linear support vector regression. b, Mean of species-wise Pearson correlation of log2-transformed MICs between values obtained experimentally and predicted by various ML models. Evaluated dataset: 69 peptides were synthesized and tested. Credit: Wan et al.
Remarkably, 13 of the 41 active molecules were classified as archaic encrypted peptides (AEPs) - sequences present in extinct but not extant organisms. These represent truly "resurrected" antimicrobials that had been lost to extinction.
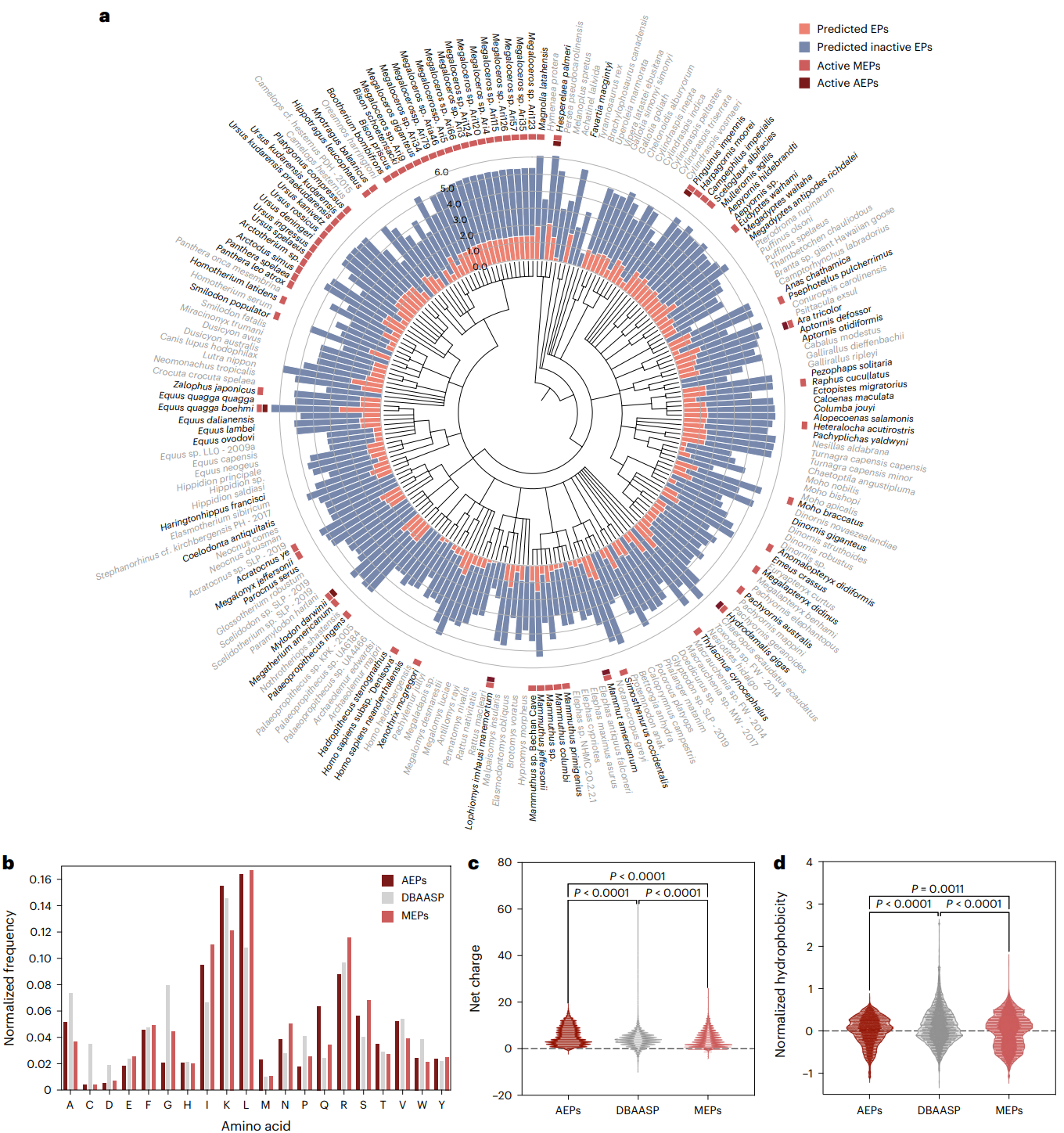
Fig. 3 | Antimicrobials identified by APEX in extinct organisms and their composition and physicochemical properties. a, Phylogenetic tree showing the extinct organisms scanned by APEX. Circular bars denote the log10- transformed average active (red) and inactive (blue) EPs discovered by APEX. A peptide was considered active when its predicted median MIC against the bacterial strains tested was ≤80 μmol l−1. The values were normalized by the number of proteins per organism scanned. The organisms whose EPs were selected for validation are highlighted in bold type. Extinct organisms that presented active EPs validated experimentally are indicated by a light red square and, within that group, those organisms encoding extinct sequences absent in extant organisms are highlighted with a dark red square. b, Amino acid frequency in AEPs and MEPs compared with known AMPs from the DBAASP database. AEPs present a higher frequency of the basic residue K, the aliphatic residue V, and uncharged polar residues (M, Q and T) than MEPs. c,d, Distribution of two physicochemical properties for peptides with predicted antimicrobial activity (AEPs and MEPs) and AMPs from DBAASP: net charge (c) and normalized hydrophobicity (d). Net charge directly influences the initial electrostatic interactions between the peptide and negatively charged bacterial membranes, and hydrophobicity directly influences the interactions of the peptide with lipids in the membrane bilayers. EPs from extinct organisms are slightly less hydrophobic and similarly have a net positive charge, compared with EPs from the modern human proteome16 or peptides from DBAASP. Statistical significance in c and d was determined using two-tailed t-tests followed by Mann–Whitney test; P values are shown in the graph. The solid line inside each box represents the mean value obtained for each group. Credit Wan et al.
The nomenclature system developed by the researchers creatively honors the source organisms: mammuthusin-2 from the woolly mammoth Mammuthus primigenius, elephasin-2 from the straight-tusked elephant Elephas antiquus, mylodonin-2 from the giant sloth Mylodon darwinii, and megalocerin-1 from the extinct giant elk Megaloceros giganteus.
Detailed mechanistic studies revealed that these ancient antimicrobials operate through a distinct mode of action compared to conventional antimicrobial peptides. Rather than primarily targeting bacterial outer membranes, the extinct-derived peptides showed exceptional ability to depolarize cytoplasmic membranes.
Using the potentiometric fluorophore DiSC₃-5, researchers demonstrated that peptides like anomalopterin-1, mylodonin-4, and equusin-2 depolarized Acinetobacter baumannii membranes more effectively than the clinical antibiotic polymyxin B. This suggests a potentially novel therapeutic mechanism that could circumvent existing resistance pathways
The physicochemical analysis revealed that AEPs possess distinct characteristics that distinguish them from both modern encrypted peptides and conventional antimicrobial peptides.
They showed higher frequencies of uncharged polar residues (methionine, glutamine, threonine) and branched aliphatic residues (isoleucine, leucine) while displaying lower amphiphilicity and reduced tendency toward aggregation. These structural features likely contribute to their unique membrane-targeting properties and may explain their effectiveness against resistant pathogens.
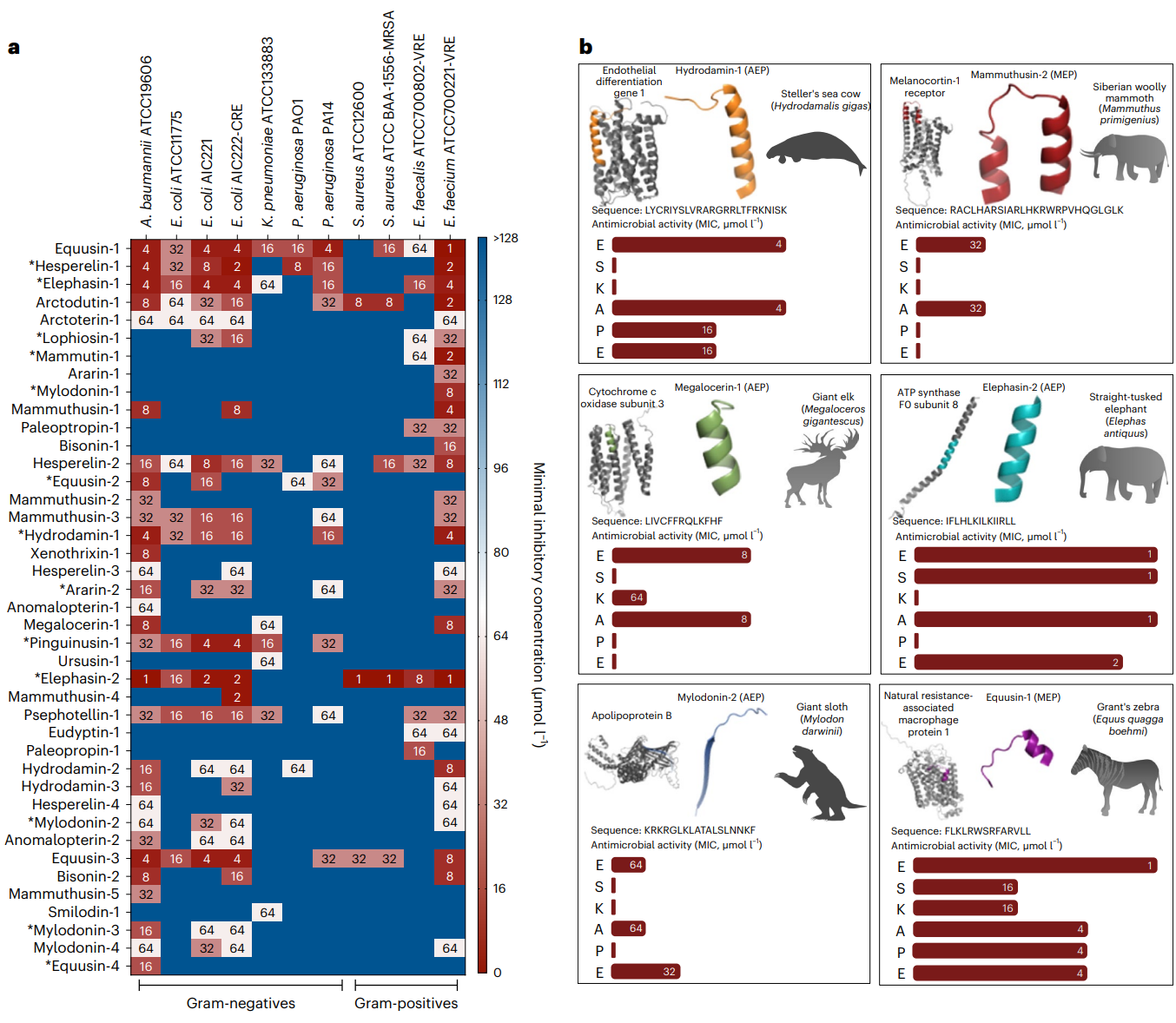
Fig. 4 | Antimicrobial activity profiles of sequences from the proteomes of extinct organisms. a, Heat map of the antimicrobial activities (μmol l−1) of the active antimicrobial agents from extinct organisms against 11 clinically relevant pathogens, including strains resistant to conventional antibiotics. Briefly, 106 bacterial cells and serially diluted EPs (0–128 μmol l−1) were incubated at 37 °C. One day post treatment, the optical density at 600 nm was measured in a microplate reader to evaluate bacterial growth in the presence of the EPs from extinct organisms. MIC values in the heat map are the arithmetic mean of the replicates in each condition. b, Examples of active AEPs and MEPs from various extinct organisms, their parent protein and their activity profile against ESKAPE pathogens (Enterococcus spp., S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, E. coli). Antimicrobial activity is expressed as the MIC (μmol l−1), and activity bars are presented as −log2 MIC14. The data for the assays in a are the mean, and the experiments were performed in three independent replicates. AEPs in a are indicated by an asterisk (*). The protein and peptide structures shown in the figure were created with PyMOL Molecular Graphics System, version 2.1 Schrödinger, LLC. Credit: Wan et al.
From Bench to Bedside: Animal Model Success
The most compelling evidence for the therapeutic potential of molecular de-extinction came from rigorous testing in two established mouse infection models. In skin abscess experiments, topical application of the antimicrobial peptides at their minimum inhibitory concentrations resulted in 2-5 order of magnitude reductions in bacterial loads compared to untreated controls. Elephasin-2 and mylodonin-2 demonstrated activity comparable to polymyxin B, a last-resort antibiotic used for multidrug-resistant gram-negative infections.
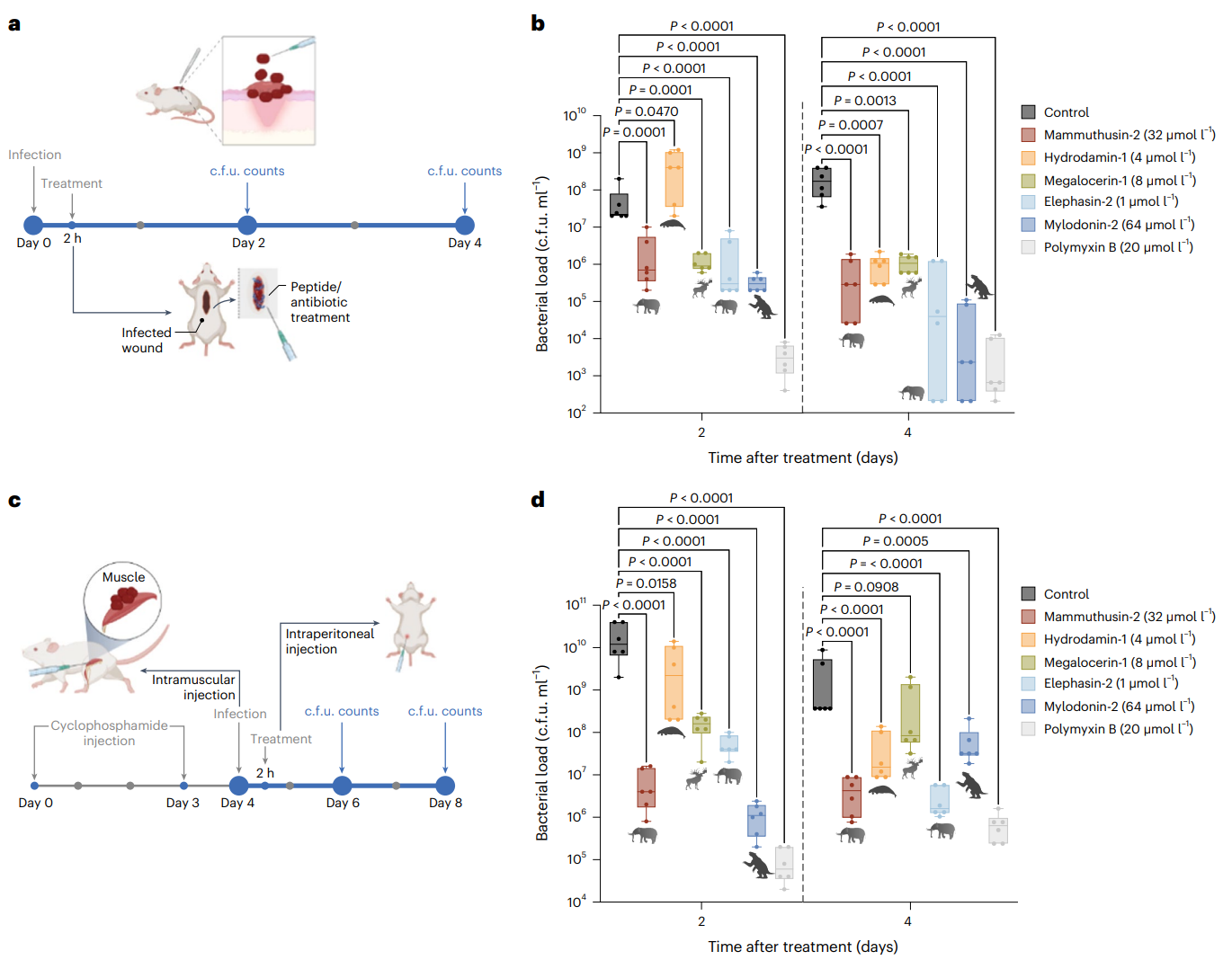
Fig. 6 | Anti-infective activity of antibiotic molecules in animal models. a, Schematic of the skin abscess mouse model used to assess the anti-infective activity of selected antimicrobials from extinct organisms (n = 6) against A. baumannii ATCC 19606. b, The molecules mammuthusin-2 (M. primigenius), hydrodamin-1 (H. gigas), megalocerin-1 (Megalocerus sp.), elephasin-2 (E. antiquus) and mylodonin-2 (M. darwinii), administered at their MIC in a single dose, inhibited the proliferation of the infection for up to 4 days after treatment compared to the untreated control group. Elephasin-2 and mylodonin-2 cleared the infection in some of the mice, with activity comparable to that of the antibiotic used as control, polymyxin B. c, Schematic of the neutropenic thigh infection mouse model in which EPs from extinct organisms were injected intraperitoneally. Anti-infective activity against A. baumannii ATCC 19606 was assessed 2 and 4 days after intraperitoneal peptide administration (n = 6). d, Two days after intraperitoneal injection, mylodonin-2 at its MIC reduced A. baumannii ATCC19606 infection as much as polymyxin B, compared to the untreated control group. Four days post treatment, mammuthusin-2 and elephasin-2 showed the same level of activity as polymyxin B. Statistical significance in b and d was determined using one-way ANOVA followed by Dunnett’s test; P values are shown in the graph. For the boxplots, the centre line represents the mean, the box limits the first and third quartiles and the whiskers (minima and maxima) 1.5 × the interquartile range. The solid line inside each box represents the mean value obtained for each group. Panels a and c were created with BioRender.com. Credit Wan et al.
The neutropenic thigh infection model provided even more stringent testing conditions, simulating the immunocompromised state of many patients with serious bacterial infections. Following intraperitoneal administration, all tested peptides except hydrodamin-1 showed significant antimicrobial efficacy within 48 hours.
Notably, mylodonin-2 from the giant sloth exhibited the most potent activity, reducing bacterial loads by 4-5 orders of magnitude - matching the performance of polymyxin B. By day four post-treatment, elephasin-2 and mammuthusin-2 had also achieved substantial bacterial clearance comparable to the control antibiotic.
Crucially, none of the tested peptides showed significant toxicity in either animal model, as evidenced by stable body weights and absence of visible tissue damage throughout the experimental periods.
Cytotoxicity assays using human embryonic kidney cells revealed that 39 of 41 active peptides showed no notable toxicity at therapeutic concentrations. The two peptides that did exhibit cytotoxicity (lophisin-1 and xenothrixin-1) still maintained therapeutic windows, with antimicrobial concentrations 8-fold lower than cytotoxic doses.
Synergy and Stability: Optimizing Ancient Arsenals
The research team discovered that combining peptides from the same extinct organism or related species often resulted in synergistic interactions that dramatically enhanced antimicrobial potency.
Checkerboard assays revealed that most tested combinations produced fractional inhibitory concentration index (FICI) values indicating synergistic or additive effects. The most remarkable example involved equusin-1 and equusin-3 from the extinct zebra Equus quagga boehmi, which reduced minimum inhibitory concentrations by 64-fold when used together, achieving sub-micromolar potency comparable to the most effective clinical antibiotics.
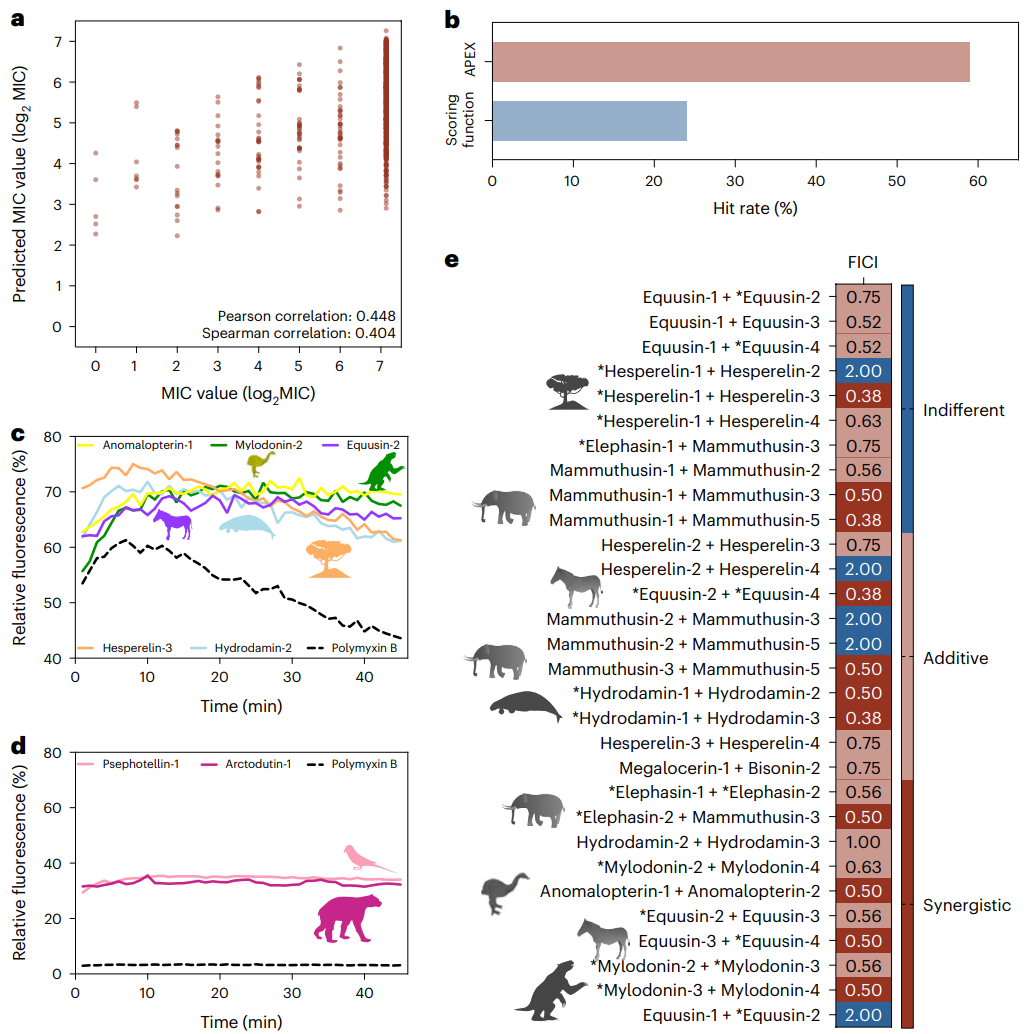
Fig. 5 | Antimicrobial activity, mechanism of action and synergy of antimicrobials from the proteomes of extinct organisms. a, Pan-bacterial Pearson and Spearman correlations of log2-transformed MICs between experimentally validated values and values predicted by APEX. b, Comparison between the hit rates of APEX and the scoring function previously described by Torres et al.16 to detect antibiotics in the modern human proteome. c, Cytoplasmic membrane depolarization by five antimicrobials from extinct organisms. The A. baumannii membrane was more strongly depolarized by the peptides than by the antibiotic polymyxin B. d, NPN permeabilization assays showing the effect of two antimicrobials from extinct organisms on the outer membrane of A. baumannii. Higher permeability was observed with the peptides than with the antibiotic polymyxin B. e, Heat map showing interactions between antimicrobials identified by APEX, expressed as the FICI. Most of the tested EP pairs from extinct organisms either synergized or had an additive effect against A. baumannii and P. aeruginosa; the latter was only tested against the peptide pair composed of equusin-1 and equusin-2 shown in the last row of the heat map. The data for the assays in c–e are the mean, and the experiments were performed in three independent replicates. AEPs in e are indicated by an asterisk (*). Credit: Wan et al.
These synergistic interactions suggest that extinct organisms may have employed cocktails of antimicrobial peptides for enhanced defense against microbial threats. Pairs showing particularly strong synergy included hesperelin-1 and hesperelin-3 from the extinct palm Hesperelaea palmeri, mammuthusin-1 and mammuthusin-3 from the woolly mammoth, and hydrodamin-1 and hydrodamin-3 from the ancient sea cow Hydrodamalis gigas. This finding has important implications for developing combination therapies that could overcome antibiotic resistance through multiple mechanisms of action.
Stability studies in human serum revealed variable proteolytic resistance among the ancient peptides. Elephasin-2 and mammuthusin-2, both derived from proboscidean species, showed the highest stability with approximately 40% remaining after six hours of exposure to human proteases.
While other peptides degraded more rapidly, their demonstrated in vivo efficacy suggests sufficient stability for therapeutic applications, particularly when formulated for localized delivery or modified to enhance protease resistance.
Future Horizons in Molecular Archaeology
The success of molecular de-extinction opens numerous avenues for future drug discovery and biological research. As ancient DNA sequencing technologies improve and more extinct proteomes become available, the molecular reservoir accessible to APEX will continue expanding.
Current limitations include the relatively small number of well-characterized extinct proteins and the focus on organisms with available genomic data. However, ongoing paleogenomics initiatives are rapidly expanding this database, with recent successes in sequencing DNA from increasingly ancient specimens.
The APEX framework could be extended beyond antimicrobial discovery to identify other bioactive molecules from extinct organisms. Anti-inflammatory compounds, enzyme inhibitors, and even novel structural proteins could be resurrected using similar computational approaches.
The method could also be applied to recently extinct species for which more complete genomic information is available, potentially yielding additional therapeutic candidates with established evolutionary validation.
Integration of structural biology approaches could further enhance the power of molecular de-extinction. While APEX currently relies solely on sequence information, incorporating predicted or experimentally determined three-dimensional structures could improve accuracy and provide insights into structure-activity relationships. This could enable rational design modifications to optimize the properties of resurrected molecules for specific therapeutic applications.
Bridging Past and Future in Drug Discovery
Molecular de-extinction represents a fundamentally new paradigm in drug discovery that leverages evolutionary history as a source of pre-validated therapeutic candidates. Unlike purely synthetic approaches that must explore vast chemical spaces without guidance, this method focuses on molecules that have already been tested by millions of years of natural selection. The success of ancient antimicrobials in modern bacterial cultures demonstrates the conservation of fundamental biological processes across deep evolutionary time.
The work also highlights the untapped potential of interdisciplinary collaboration between computational biology, paleogenomics, and pharmaceutical science. By combining cutting-edge machine learning algorithms with paleobiological data, the researchers have created a new field of computational archaeology that could revolutionize our approach to therapeutic development. This integration of historical and technological perspectives offers a unique solution to the urgent challenge of antibiotic resistance.
As we face an uncertain future where established antibiotics lose effectiveness against evolving pathogens, the molecular wisdom preserved in extinction offers hope. The APEX algorithm and the concept of molecular de-extinction provide both immediate therapeutic candidates and a roadmap for accessing the vast pharmacological heritage encoded in the tree of life. By learning from the past, we can better prepare for the microbial challenges of the future, ensuring that the evolutionary innovations of extinct species continue to protect life on Earth.
Definitions
APEX (Antibiotic Peptide de-Extinction): A deep learning algorithm that combines recurrent neural networks with attention mechanisms to predict antimicrobial activity of peptides from extinct organisms.
Archaic Encrypted Peptides (AEPs): Antimicrobial peptide sequences found in extinct organisms but absent from modern species, representing truly "resurrected" molecules.
Cytoplasmic Membrane Depolarization: The disruption of the electrical potential across bacterial cell membranes, leading to cell death through metabolic collapse.
Encrypted Peptides (EPs): Short amino acid sequences within larger proteins that possess antimicrobial properties when isolated and tested independently.
ESKAPEE Pathogens: A group of dangerous bacterial species (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli) identified by WHO as priority threats.
Fractional Inhibitory Concentration Index (FICI): A measure of antimicrobial interaction, where values ≤0.5 indicate synergy, 0.5-4.0 indicate additivity or indifference, and >4.0 indicate antagonism.
Minimum Inhibitory Concentration (MIC): The lowest concentration of an antimicrobial agent that prevents visible bacterial growth after overnight incubation.
Molecular De-extinction: The computational resurrection of bioactive molecules from extinct organisms to address contemporary biological and medical challenges.
Multitask Learning: A machine learning approach that simultaneously optimizes multiple related objectives, in this case predicting both antimicrobial activity and binary classification of peptides.
Uniform Manifold Approximation and Projection (UMAP): A dimensionality reduction technique used to visualize high-dimensional sequence data in two-dimensional space for comparative analysis.

Deep Learning Resurrects Ancient Antibiotics: Mining Extinct Proteomes for Modern Medicine
Deep-learning-enabled antibiotic discovery through molecular de-extinction