Designing DNA and RNA with precision is crucial for advances in modern therapeutics, but the vastness of biological sequence space makes this an immense computational challenge. Traditional search methods can't keep pace, but recent breakthroughs in artificial intelligence are reshaping what's possible.
Google's NucleoBench benchmark and the AdaBeam algorithm are at the forefront, enabling smarter, faster, and more scalable nucleic acid sequence design for drug discovery and beyond.
The Multifaceted Challenge of Sequence Design
Crafting effective nucleic acid sequences involves several complex steps:
- Data generation: Collecting examples with desired biological properties
- Model training: Using AI to predict outcomes for new sequences
- Sequence generation: Optimizing algorithms to propose novel candidates
- Experimental validation: Testing the most promising designs in the lab
The optimization stage is especially daunting due to the sheer number of possibilities and lack of standard benchmarks, making it difficult to compare algorithm performance.
NucleoBench: Leveling the Playing Field
NucleoBench addresses this inequality by offering a transparent, rigorous framework for algorithm evaluation. Covering 16 biologically relevant tasks and over 400,000 experiments, it compares both gradient-free (like simulated annealing and directed evolution) and gradient-based (such as FastSeqProp and Ledidi) algorithms. Tasks span gene expression control, transcription factor binding, chromatin accessibility, and sequence design for large-scale models, bringing much-needed clarity to the field.
AdaBeam: Next-Generation Sequence Optimization
The standout innovation, AdaBeam, is a hybrid adaptive beam search algorithm. It merges the diversity-focused approach of unordered beam search with the efficient expansion of AdaLead, maintaining a pool of high-potential sequences and expanding on the best candidates. Its advantages include:
- Efficiency: Rapidly samples new candidates, doubling the speed on longer sequences
- Smart exploration: Flexibly chooses which sequence positions to edit for optimal results
- Scalability: Uses gradient concatenation to reduce memory usage, making it suitable for large models
AdaBeam's adaptive strategy lets it focus on promising regions within the sequence landscape, excelling where traditional algorithms struggle.
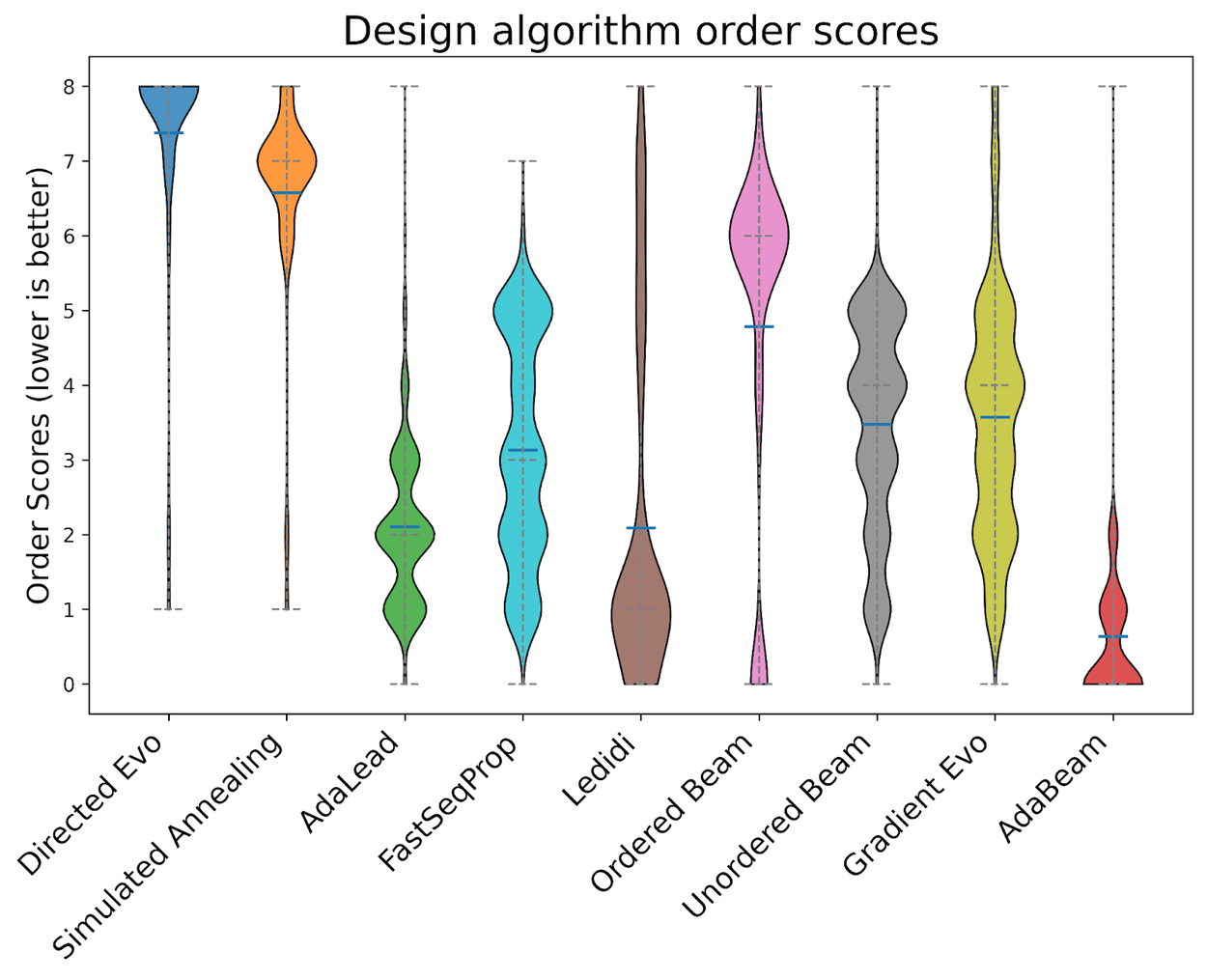
The distribution of final scores for each algorithm. X-axis is the design algorithm, y-axis is the aggregate order score. Order scores are determined by assigning an integer [0, 9] for each (task, start sequence, design algorithm) tuple according to the performances of all the final sequences for that (task, start sequence) pair. 0 is the top performer. Aggregate scores are computed by averaging over all such scores. Credit: Google Research
Benchmarking Results and Insights
All nine algorithms, including AdaBeam, were tested under fair, controlled conditions: identical starting sequences, equal computational resources, and multiple random seeds to ensure robust results.
Evaluation metrics included fitness scores, speed of convergence, and resilience to initial conditions. AdaBeam surpassed its peers in 11 of 16 tasks, offering faster convergence and exceptional scalability.
The study also highlighted that:
- Gradient-based algorithms are insightful but can be slow and memory-intensive
- AdaBeam balances speed, scalability, and effectiveness, particularly for long, complex sequences
Additionally, the choice of starting sequence was shown to have a significant impact on results, and not all classic algorithm features transfer seamlessly to biological design.
Open Tools, Responsible Innovation
By releasing NucleoBench and AdaBeam as open-source resources, Google and Move37 Labs invite the community to iterate, innovate, and set new benchmarks. This approach fosters transparency, accelerates progress, and ensures that algorithmic advances align with real biological constraints. Importantly, AdaBeam works within the safety boundaries of predictive models and is designed for responsible use, requiring expert oversight.
A Smarter Path Forward
NucleoBench and AdaBeam establish new best practices for nucleic acid design, marrying rigorous benchmarking with cutting-edge optimization. These tools are poised to hasten the development of transformative therapies, from vaccines to gene editing, by making the design process more efficient and accessible. The journey ahead promises even greater collaboration and breakthroughs in AI-powered biological discovery.
Source: Google Research Blog

Smarter Nucleic Acid Design: How NucleoBench and AdaBeam Are Unlocking the Future of Nucleic Acid Engineering