The rapid identification of pathogenic bacteria is of paramount importance in the effective management of infectious diseases, particularly in acute and life-threatening conditions such as sepsis. Sepsis, currently the 3rd leading cause of deaths in the US is a systemic inflammatory response to infection that can rapidly progress to organ dysfunction and death if not promptly diagnosed and treated. Early identification of the causative pathogen allows for targeted antimicrobial therapy, which is crucial in improving patient outcomes and reducing mortality rates.
Traditional methods for microbial identification, such as culture and biochemical testing, can be time-consuming, therefore, there is a growing need for rapid diagnostic techniques that can accurately identify pathogens within a matter of hours, enabling clinicians to make informed treatment decisions and optimize patient care.
This research presents a novel approach for the rapid and accurate identification of multiple bacterial species directly from samples, bypassing the need for time-consuming culturing or sequencing methods. The study leverages peptide nucleic acid (PNA) probes in conjunction with fluorescence in situ hybridization (FISH) and advanced techniques like Förster resonance energy transfer (FRET) and chemically cleavable fluorophores to achieve high specificity and speed in microbial detection.
Key Takeaways
- Species-specific PNA probes designed using 16S rRNA sequence variation analysis achieved 96-99.9% accuracy in identifying seven bacterial species commonly found in bacteremia.
- PNA probes demonstrated superior penetration into bacteria and higher mismatch sensitivity compared to traditional DNA probes, leading to more efficient and specific target detection.
- FRET-based FISH utilizing pairs of adjacently binding PNA probes virtually eliminated inter-species crosstalk, significantly enhancing identification accuracy.
- A rapid sequential species identification method was implemented using chemically cleavable fluorophores linked to PNA probes, allowing for the identification of multiple species in a mixture without compromising accuracy and significantly reducing processing time compared to photobleaching.
- The developed techniques hold high potential for clinical applications, particularly in the timely diagnosis and treatment of acute infectious diseases like sepsis and bacteremia.
Overview
The accurate and rapid identification of pathogenic microbes is critical for the effective treatment of infectious diseases, especially acute conditions like sepsis. Current gold standard methods like PCR amplification and MALDI-TOF mass spectrometry have significant limitations in time and potential for error.
PCR often requires a culturing step due to its detection limits being higher than typical bloodstream infection concentrations. While MALDI-TOF struggles with identifying species in mixed samples and also frequently necessitates prior colony culture. These limitations highlight the demand for faster, unbiased, and more accurate diagnostic alternatives.
Fluorescence in situ hybridization (FISH) has emerged as a promising culture-independent technique for microbial identification. FISH utilizes fluorescently labeled oligonucleotide probes that hybridize to specific ribosomal RNA (rRNA) sequences within microbial cells.
The variability in rRNA sequences, particularly the 16S rRNA in bacteria, makes it a suitable target for distinguishing between species. However, traditional DNA-based FISH probes face challenges related to bacterial cell wall permeability and limited sensitivity to mismatches, which can hinder their effectiveness in complex microbial mixtures.
To overcome these limitations, the researchers in this study focused on utilizing peptide nucleic acids (PNAs) as the backbone for their FISH probes. PNAs are synthetic DNA analogs where the sugar-phosphate backbone is replaced by a charge-neutral pseudo-peptide structure. This unique characteristic of PNAs offers several advantages over DNA probes for bacterial FISH.
Their charge neutrality enhances hybridization stability by minimizing electrostatic repulsion. This promotes better penetration through bacterial membranes and reduces hybridization time. PNA probes also exhibit higher sensitivity to even single base mismatches, enabling improved discrimination between closely related bacterial species. The research team hypothesized that PNA-based FISH, with optimized protocols, could provide a fast and accurate solution for multi-bacterial identification.
Why it’s Important
The advancements presented in this research have significant implications for clinical microbiology and the diagnosis of infectious diseases. The ability to rapidly and accurately identify bacterial pathogens directly from patient samples, without the need for lengthy culturing steps, can dramatically improve the timeliness and effectiveness of treatment, especially in life-threatening conditions like bacteremia and sepsis.
Early and targeted antibiotic therapy, guided by precise pathogen identification, can lead to better patient outcomes and potentially reduce the development of antibiotic resistance associated with broad-spectrum treatments.
The development of PNA-based FISH with enhanced specificity through FRET further addresses the challenge of false positive signals and crosstalk in complex microbial environments. This increased accuracy is crucial for reliable diagnostic assays.
Moreover, the introduction of chemically cleavable fluorophores for sequential FISH opens the door for rapid and multiplexed detection of multiple pathogens in a single sample, streamlining the diagnostic workflow and potentially reducing costs and turnaround times.
This approach could be particularly valuable in clinical settings where polymicrobial infections are common. The research highlights a promising alternative to existing molecular diagnostic methods, offering a culture-free and highly specific imaging-based approach.
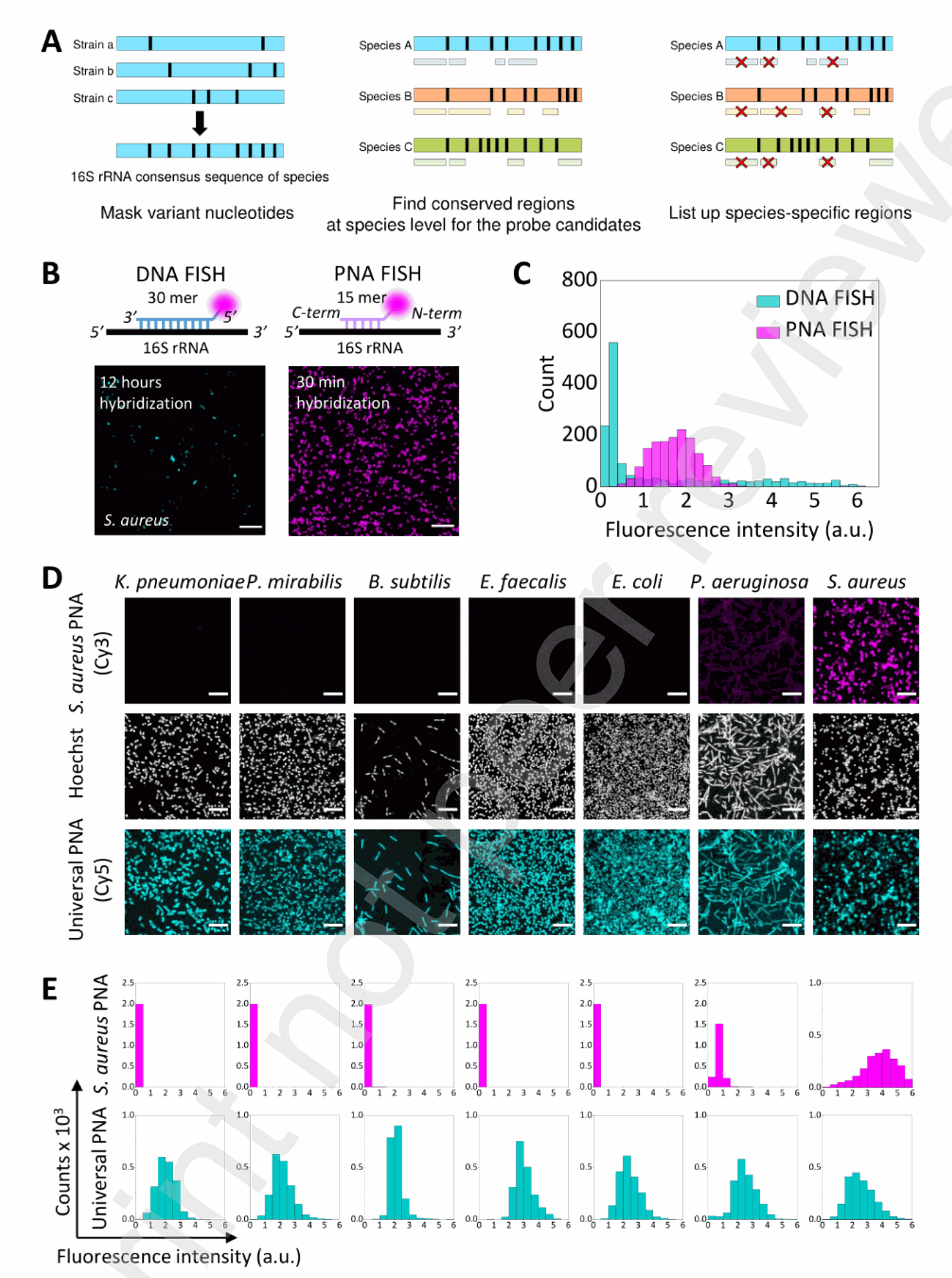
Figure 1. Design and characterization of PNA probes for bacterial FISH (A) The design principle of species-specific FISH probes developed in this study. Black stripes show the regions variable between strains. Consensus regions unique to each species were chosen for probe design. (B) Schematic comparison of the DNA and PNA probes showing the label position (magenta circles) and the results of S. aureus imaging by their respective FISH protocols. Scale bar: 15 μM. (C) Histograms of mean fluorescence signal of bacterial cells from Cy3-labeled DNA and PNA FISH images, shown in cyan and magenta, respectively (n = 1,500 for each histogram). Fluorescence intensity is shown in an arbitrary unit, which is consistent throughout the study for fair comparison. (D) Fluorescence images of seven bacterial species hybridized with Cy3-labeled SAUR.69 (magenta) and Cy5-labeled UNIV (cyan) probes, shown along with Hoechst images (greyscale) of the same region. Scale bar: 10 μM. (E) Histograms of the fluorescence signal of SAUR.69 (magenta) and UNIV (cyan) probes in the same order as in (D) (n = 2,000 for each histogram).
Summary of Results
The researchers began by designing a set of 15-base species-specific PNA probes targeting the 16S rRNA of seven bacterial species commonly found in bacteremia: Klebsiella pneumoniae, Proteus mirabilis, Bacillus subtilis, Enterococcus faecalis, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus.
The probe sequences were carefully selected by analyzing variations in the 16S rRNA database to maximize mismatches with non-target species (Fig. 1A, Table 1). For comparison, a longer 30-mer DNA probe targeting S. aureus was also designed.
The PNA FISH procedure was rigorously optimized by testing various permeabilization reagents, hybridization times, formamide concentrations, and washing conditions (Sup. Fig. S2, S3, S4). The optimized protocol involved permeabilization with lysozyme, hybridization at 55°C with 30% formamide, and washing with 15% formamide.
Comparative analysis revealed that the PNA probe (SAUR.69) targeting S. aureus exhibited significantly stronger and more homogeneous fluorescence signals compared to the DNA probe (SAUR.211 (DNA)), demonstrating superior target binding efficiency and cell penetration (Fig. 1B, C). Specificity testing showed that each PNA probe displayed minimal crosstalk with non-target species, with most combinations exhibiting >99% specificity (Fig. 1D, E, Fig. 2A, B). The only notable crosstalk (3.7%) was observed between the S. aureus probe (SAUR.69) and P. aeruginosa.
To further enhance specificity and eliminate potential crosstalk, a FRET-based FISH assay was developed using pairs of PNA probes targeting adjacent regions on the 16S rRNA (Fig. 3A).
When both a donor (Cy3-labeled) and an acceptor (Cy5-labeled) probe bind adjacent sites on the target rRNA, excitation of the donor leads to energy transfer and emission from the acceptor, generating a FRET signal.
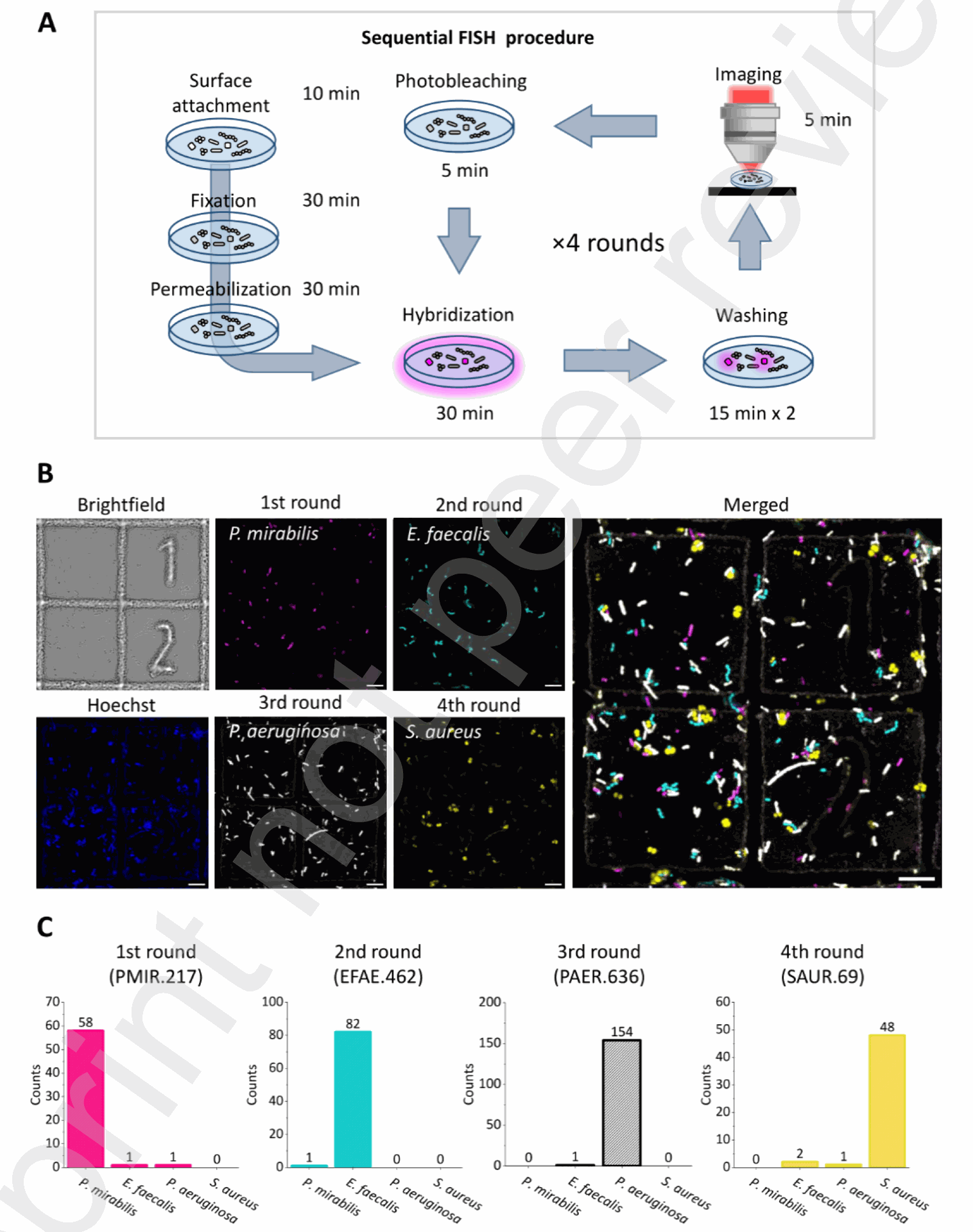
Figure 4. Sequential FISH of multi-species mixture with PNA probes (A) Schematic procedure of sequential FISH with PNA probes. After surface attachment, fixation, and permeabilization of bacterial cells, cycles of hybridization, washing, imaging, and photobleaching followed. The time required for each step is shown. (B) Brightfield, Hoechst, and FISH images of the bacterial mixture with PNA probes targeting P. mirabilis (magenta), E. faecalis (cyan), P. aeruginosa (white), and S. aureus (yellow), shown along with a merged image. Scale bars: 10 μm. (C) Counts of bacterial cells detected in each round of FISH imaging. Spots overlapping in multiple FISH images were counted as potential crosstalk. Further analyses on the overlapping spots are in Sup. Fig. S7. Experiments with E. coli-specific probe pairs demonstrated a high FRET efficiency (0.8) in the target species, while non-target species showed negligible FRET signals (Fig. 3B-E). This FRET-based detection significantly improved the separation between target and non-target species compared to using a single probe (Fig. 2A, 3E). Control experiments with probes targeting distal sites or the same site on different rRNA molecules confirmed that a strong FRET signal requires adjacent binding on the same target molecule (Fig. 3F-I).
The researchers also demonstrated sequential identification of multiple bacterial species in a mixture using repeated rounds of PNA FISH with different Cy3-labeled probes, followed by photobleaching between rounds (Fig. 4A, B).
Analysis of fluorescence signals from individual cells across the sequential images showed minimal crosstalk between the different detection rounds (Fig. 4C, Sup. Fig. S6, S7). To expedite the sequential FISH process, chemically cleavable PNA probes were developed, where the fluorophore was linked via a disulfide bond.
Treatment with TCEP rapidly cleaved the disulfide bond, removing the fluorescent signal without destabilizing the bound PNA probe (Fig. 5A-D, Sup. Fig. S8). Sequential FISH using these cleavable probes on a mixture of E. coli and S. aureus showed no noticeable crosstalk and significantly reduced the time required between detection rounds (Fig. 5E-H).
Conclusion
This research presents a significant advancement in FISH-based bacterial identification through the utilization of PNA probes and innovative techniques like FRET and chemical cleavage. The developed PNA probes exhibit superior performance in terms of specificity, penetration, and target binding compared to traditional DNA probes. The FRET-based approach further enhances identification accuracy by minimizing non-specific binding.
The implementation of chemically cleavable probes enables rapid sequential detection of multiple species in a mixture, paving the way for more efficient and comprehensive microbial diagnostics. These advancements hold considerable promise for improving the diagnosis and management of acute bacterial infections.

Fast and Accurate Multi-Bacterial Identification Using Advanced Nucleic Acid Probes