The scientific discovery is a complex, iterative process involving rigorous background research, the formulation of hypotheses, the design and execution of experiments, and the careful analysis and interpretation of resulting data.
Despite significant advancements in applying artificial intelligence (AI) to various stages of this process, automating the entire workflow from hypothesis generation to experimental data analysis within a single system has remained a challenge.
The manual synthesis of ever-growing scientific knowledge poses a significant bottleneck, particularly in fields like drug development, which require integrating diverse biological, clinical, and pharmaceutical expertise.
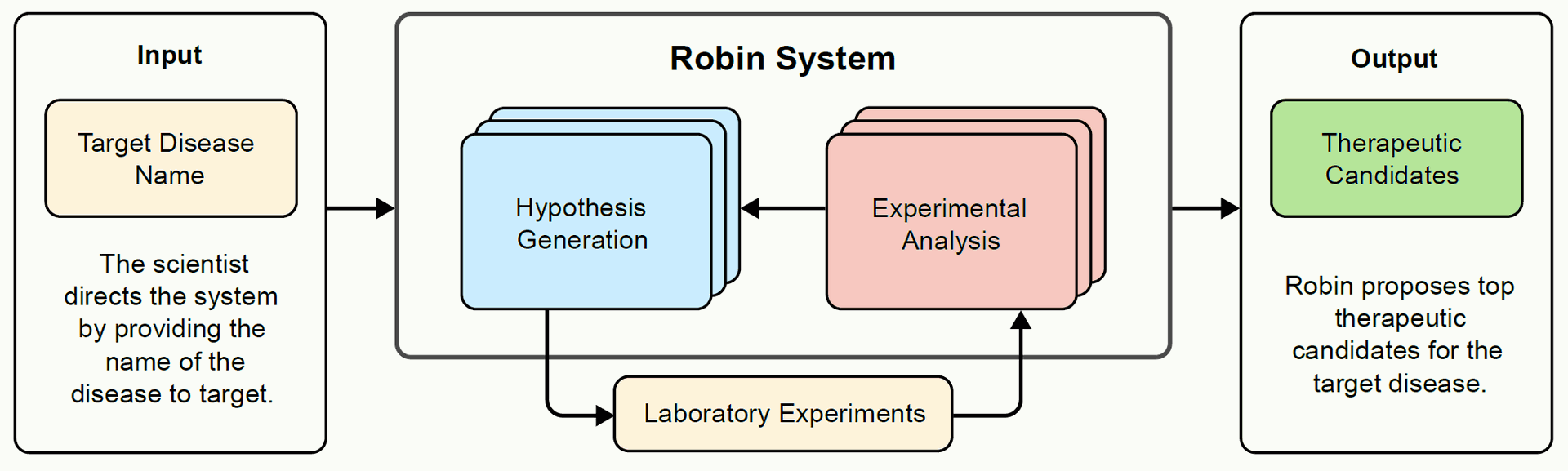
This research introduces Robin, a novel multi-agent AI system designed to automate the key intellectual steps of scientific discovery, integrating hypothesis generation with experimental data analysis in a continuous workflow.
Before Robin, FutureHouse developed several task-specific AI agents: Crow, Falcon, and Owl for deep literature review, Phoenix for synthesis design, and Finch for data analysis.
Each excelled in isolation, but integrating their expertise was the real breakthrough. Robin orchestrates these agents, automating the entire pipeline from hypothesis generation through data validation, fundamentally changing the approach to research.
By doing so, Robin aims to accelerate the pace of discovery, exemplified here by its application to finding new therapeutic candidates for dry age-related macular degeneration (dAMD).
Remarkably, Robin handled all hypothesis generation, experimental planning, data analysis, and even produced the main manuscript figures. Human scientists performed only the physical experiments, while the entire intellectual process was managed by AI.
Key Takeaways
- Full Scientific Workflow Automation: Robin is presented as the first multi-agent system capable of automating the core intellectual steps of scientific discovery, including hypothesis generation, proposing experiments, interpreting results, and refining hypotheses.
- Integration of Agents: The system integrates specialized language agents like Crow and Falcon for literature search and Finch for experimental data analysis, enabling a semi-autonomous approach.
- Hypothesis Generation: Crow reviewed scientific literature and hypothesized that boosting retinal pigment epithelium (RPE) phagocytosis could help patients with dry age-related macular degeneration (dAMD).
- Novel Therapeutic Discovery: Applying Robin to dry age-related macular degeneration (dAMD) led to the identification and validation of ripasudil, a clinically used ROCK inhibitor, as a promising therapeutic candidate that had not been previously proposed for dAMD treatment for its effect on phagocytosis.
- Mechanistic Insight Generation: Through automated RNA-seq analysis by Finch, Robin uncovered potential molecular mechanisms underlying the therapeutic effect, specifically the upregulation of ABCA1, a critical lipid efflux pump and possible novel target.
- Iterative Lab-in-the-Loop: Robin facilitates an iterative cycle where hypotheses are generated, tested experimentally (by human scientists), analyzed autonomously by the system, and then refined based on the experimental results.
- Iterative Discovery: Leveraging previous findings, Robin also proposed new candidates, leading to the identification of ripasudil, a glaucoma drug, as a novel therapy candidate for dAMD.
- Accelerating Drug Repurposing: The system's ability to synthesize disparate literature holds significant potential for identifying new uses for existing drugs, potentially overcoming the historical delays often seen in drug repurposing.
Overview
Scientific discovery relies on a cyclic process that begins with understanding existing knowledge (background research), forming potential explanations or solutions (hypothesis generation), testing these ideas through controlled observations or manipulations (experimentation), and making sense of the collected information (data analysis).
The sheer volume of scientific literature has exploded due to advances in measurement and modeling technologies, making it increasingly difficult for individual researchers to synthesize all relevant information manually.
Large language models (LLMs), trained on enormous diverse datasets, offer a potential solution by their ability to store, recall, and synthesize information across fields. They show promise in automating knowledge synthesis and accelerating scientific discovery.
One area with significant potential is drug development, particularly drug repurposing, which involves finding new uses for existing approved drugs. Historically, insights for repurposing have often existed in the literature for years before a new treatment application was realized, as seen with examples like dabrafenib (10-year lag) and ketamine (22-year lag).
This highlights the challenge of connecting disparate pieces of knowledge by human researchers. While previous AI systems have tackled specific aspects like hypothesis generation or predicting drug properties, none had integrated all key intellectual steps of the scientific process into a single, automated workflow until Robin.
Robin addresses this gap by being the first multi-agent system to combine novel hypothesis generation with experimental data analysis in a continuous process, enabling semi-autonomous scientific discovery.
The system operates through specialized language agents:
- Crow: Conducts concise literature searches to identify experimental strategies and therapeutic candidates.
- Falcon: Performs deep literature reviews to generate comprehensive reports evaluating therapeutic candidates.
- Finch: Analyzes experimental data, such as flow cytometry and RNA-sequencing.
Given a disease name, Robin uses these agents to iteratively generate hypotheses, propose experiments, analyze the resulting data, and refine its understanding to propose updated therapeutic candidates. This forms an "iterative lab-in-the-loop" framework, coordinating with human scientists who perform the physical experiments.
Why it’s Important
The development of new therapeutics is a lengthy, costly, and often inefficient process. The historical delays in drug repurposing, where known mechanisms or compounds take years to be applied to new indications despite existing literature, underscore the need for better methods to review scientific knowledge.
With the rate of FDA approvals for novel drugs holding steady at around 50 per year over the last decade, new approaches are urgently needed to accelerate discovery.
Robin represents a significant step towards addressing this need by automating the core intellectual functions of the scientific method. By rapidly sifting through and synthesizing vast amounts of literature, generating hypotheses, and autonomously interpreting complex experimental data, Robin can potentially accelerate the pace of discovery compared to traditional, entirely manual approaches.
Its ability to integrate literature insights with experimental results in a feedback loop, allows for data-driven refinement of hypotheses, which is crucial for efficient exploration of potential therapies. The identification of ripasudil for dAMD, a novel application for an existing ocular drug, demonstrates the potential for AI systems to uncover promising drug repurposing opportunities that might otherwise be missed or significantly delayed.
The system's capacity to propose and analyze mechanistic follow-up experiments, like the RNA-seq study which revealed ABCA1 upregulation, highlights its potential not just for finding treatments but also for generating deeper biological insights into disease pathways.
Beyond the findings presented in the research, there are potentially broader implications for the scientific community. Automating the iterative scientific process could free up human scientists from tedious literature review and initial data analysis, allowing them to focus on experimental design, validation, and higher-level strategic thinking.
While demonstrated in therapeutic discovery, the framework of literature synthesis, hypothesis generation, experimental design suggestion, and data analysis could potentially be applied to accelerate discovery in other scientific domains facing data overload, such as materials science, environmental science, or fundamental biology.
The development also raises questions about how best to validate AI-generated hypotheses and ensure the interpretability and reproducibility of AI-driven data analysis, areas that will require continued attention as these systems evolve.
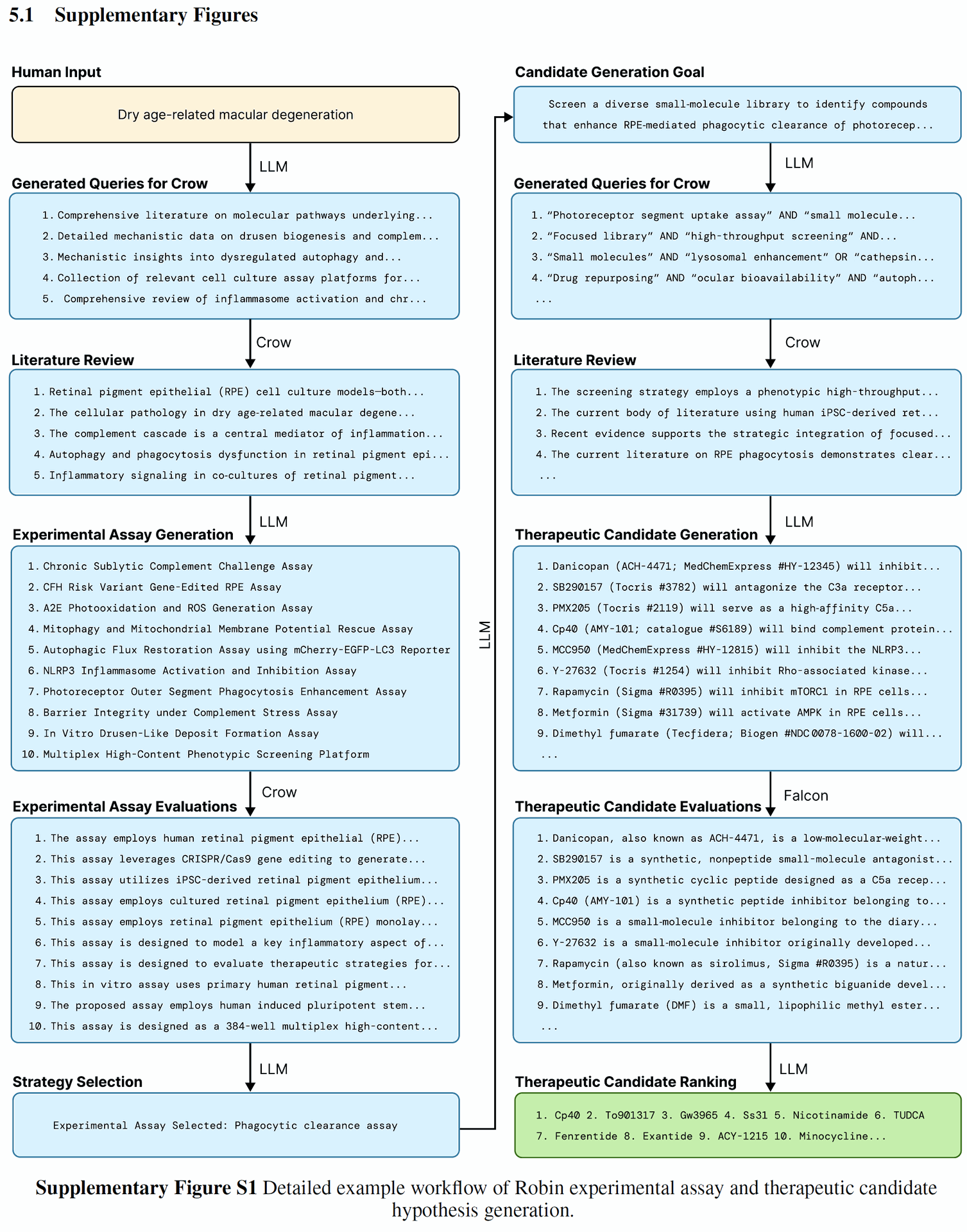
Summary of Results
The researchers applied Robin to the challenge of identifying novel therapeutic candidates for dry age-related macular degeneration (dAMD), a major cause of vision loss with limited treatment options.
The first cycle of discovery involved Robin generating initial therapeutic hypotheses:
- Given the disease name (dry age-related macular degeneration), Robin first formulated questions and used the Crow agent to review literature about dAMD pathology.
- Based on this review, Robin identified 10 potential causal disease mechanisms and used Crow to generate detailed reports on in vitro models and assays for each.
- Using an LLM judge, Robin ranked these potential strategies. The top-ranked strategy proposed treating dAMD by increasing the phagocytosis (engulfment) of material by retinal pigment epithelium (RPE) cells, and suggested testing drug efficacy in a flow cytometry assay using RPE cells.
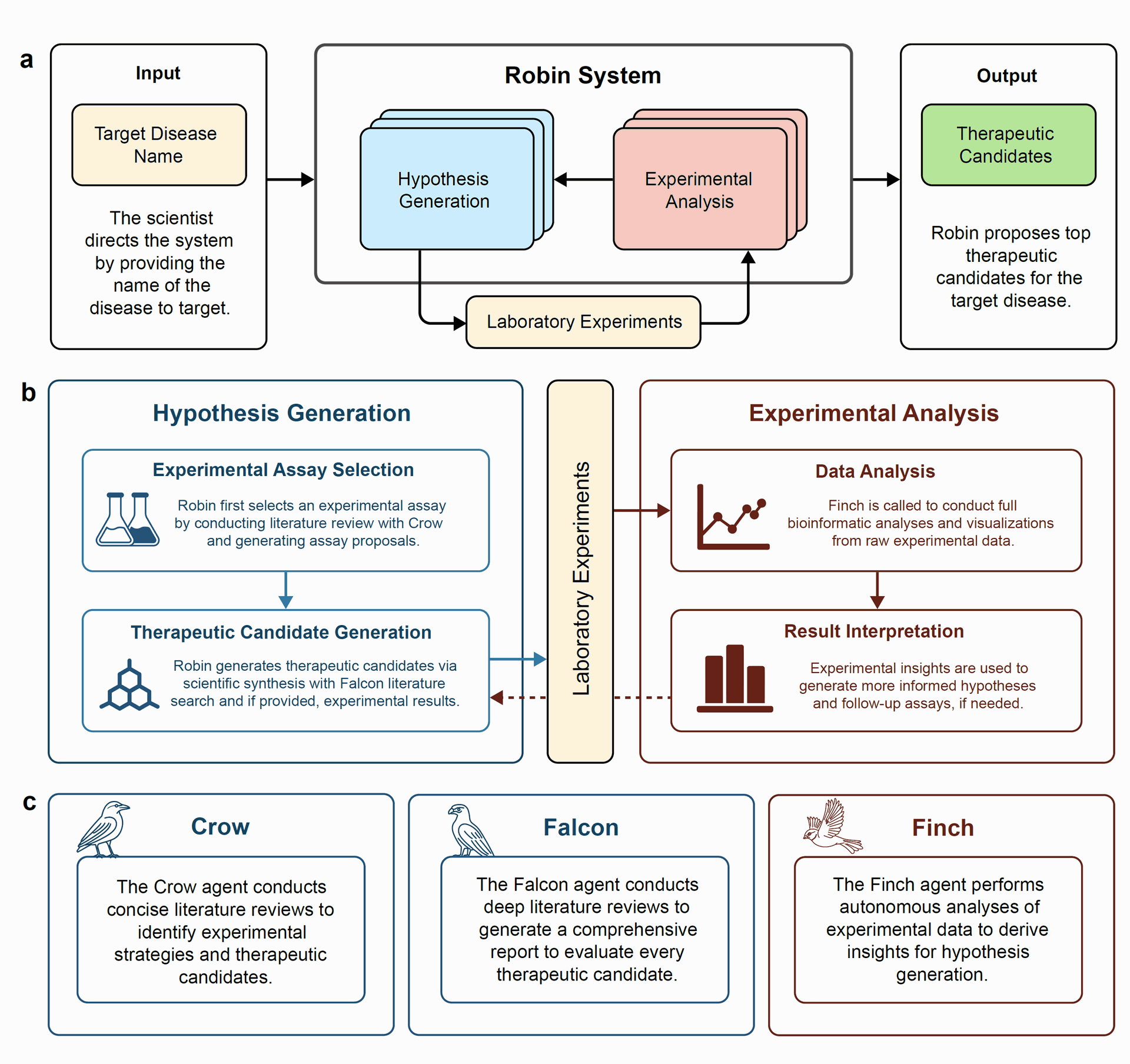
Figure 1: Architecture and workflow of the Robin system. A) Given the name of a target disease, Robin generates hypotheses and selects top therapeutic candidates to test experimentally. Robin can autonomously analyze raw data from these experiments to synthesize scientific insights and generate updated therapeutic hypotheses.
Figure 1A illustrates this initial phase, showing the input (target disease name) and output (therapeutic candidates) of the Robin system. Figure 2A shows Robin proposing several experimental assays and selecting the RPE phagocytosis enhancement assay as the strategy.
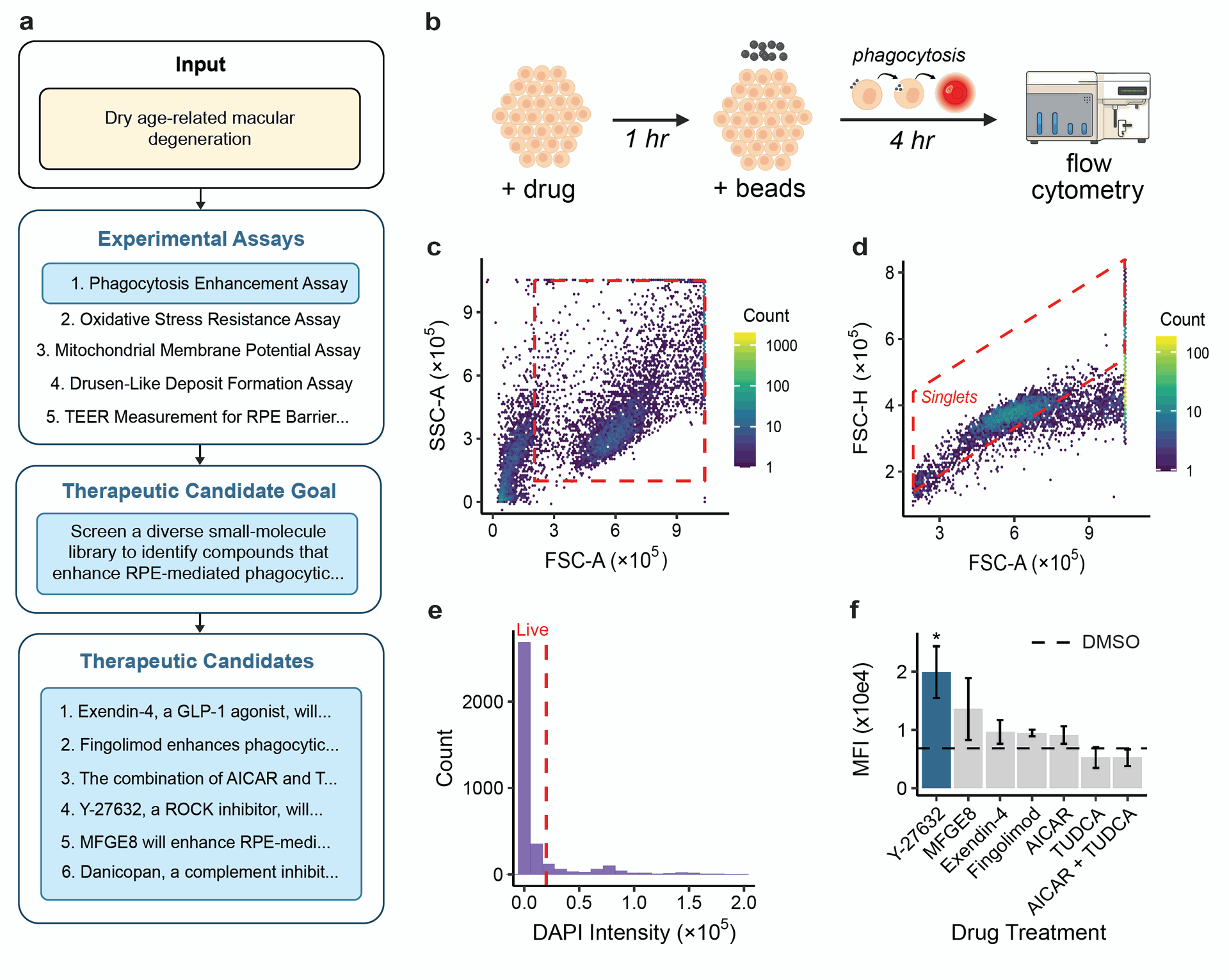
Figure 2: Robin generates therapeutic candidate hypotheses for dAMD and analyzes experimental data from in vitro tests. A) Robin proposes several experimental assays and ultimately decides to use an RPE phagocytosis enhancement assay. Robin synthesizes this strategy into an overall goal and then generates several novel therapeutic candidates to enhance RPE phagocytosis.
- Once the in vitro model was selected, Robin reviewed an additional ~400 papers related to RPE phagocytosis and dAMD therapeutics.
- It then proposed 30 therapeutic candidates for experimental testing by conducting a literature review with Crow.
- The Falcon agent was then used to create comprehensive evaluation reports for each candidate, which were ranked by an LLM-judged tournament based on scientific rationale, pharmacological profile, and supporting literature.
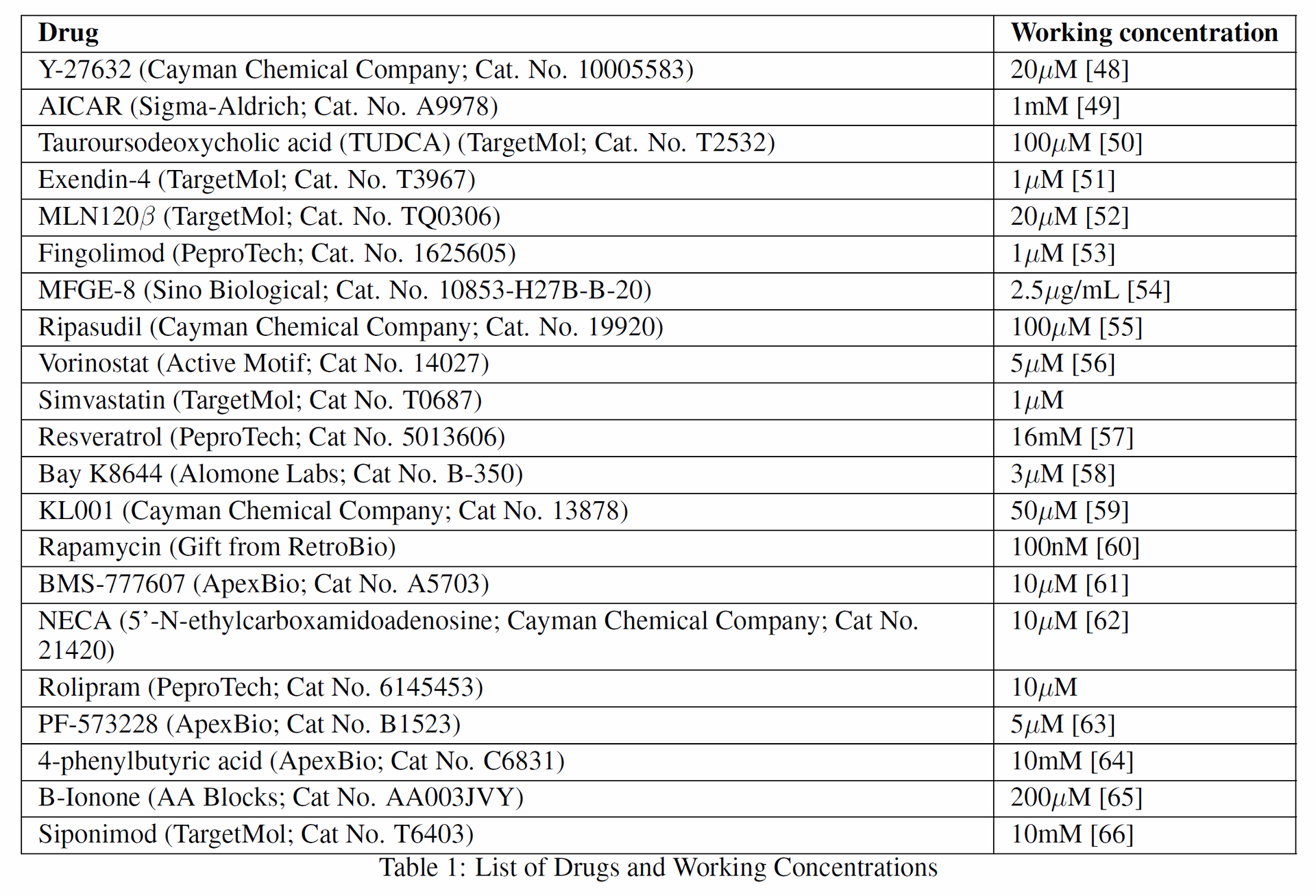
Table 1: List of Drugs and Working Concentrations [See source for Table 1, listing 22 drugs including Y-27632, AICAR, TUDCA, Exendin-4, Ripasudil, etc., and their concentrations. Table 1 lists the drugs and their working concentrations used in the subsequent experimental testing.
The experimental phase and data analysis:
- Human researchers selected the top five candidates from Robin's ranked list for experimental testing: Exendin-4, Fingolimod, MFGE8, Y-27632, and the combination of AICAR and TUDCA.
- The experiments were conducted using an RPE phagocytosis assay with pHrodo beads and flow cytometry. pHrodo beads fluoresce in the acidic environment of lysosomes, allowing quantification of engulfed material by flow cytometry. Figure 2B provides a schematic of this assay.
- Raw flow cytometry data was provided to Robin, which deployed the Finch agent for autonomous analysis.
- Finch developed a Jupyter notebook script to quantify the effect of each compound on RPE phagocytosis.
- The analysis involved gating, a process in flow cytometry where specific cell populations (e.g., live, single cells) are isolated based on their characteristics using plots like forward scatter vs. side scatter or pulse width vs. area. Finch then performed statistical tests to compare drug treatments to controls and plotted the results.
- These results were confirmed by a human scientist's analysis, which followed a similar gating strategy and statistical approach. Preclinical studies had previously shown Y-27632 could restore RPE phagocytic efficiency, supporting Robin's initial literature-based hypothesis.
The second cycle involved mechanistic investigation via RNA-seq:
- Based on the initial results, Robin recommended a follow-up RNA-sequencing experiment (a technique providing insight into gene expression) on Y-27632-treated RPE cells to understand the transcriptional effects of ROCK inhibition.
- Human researchers conducted the experiment, and Finch performed the data analysis.
- Finch conducted differential gene expression (DGE) analysis, which identifies genes whose expression levels change significantly between different conditions (e.g., treated vs. untreated cells).
- Finch performed GO enrichment analysis, which identifies biological pathways or functions that are statistically over-represented among the differentially expressed genes (Figure 3D).
- This analysis revealed changes in genes related to actin filament organization, small GTPase signaling, and autophagy pathways.A human analysis also confirmed similar DGE and GO enrichment results.
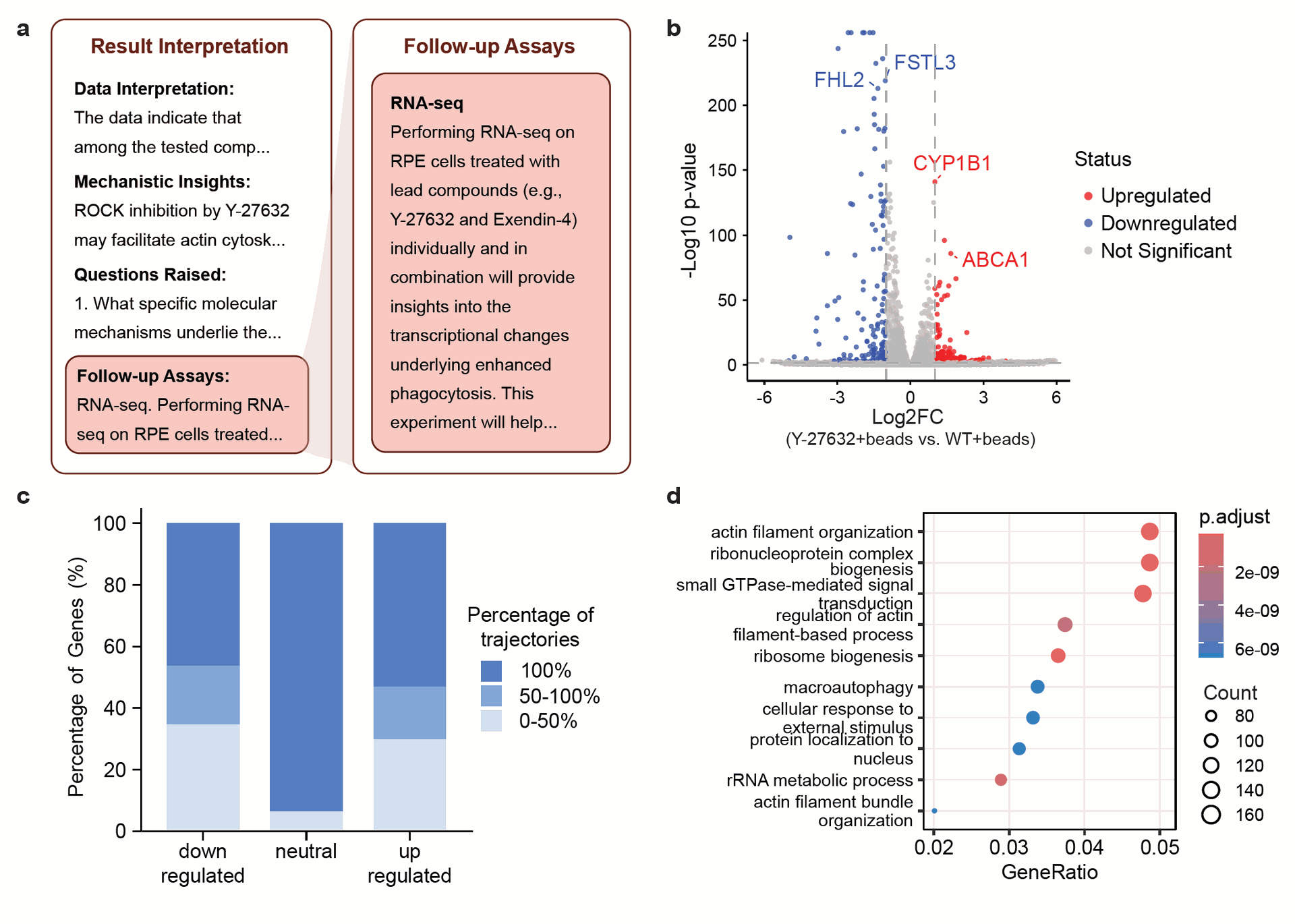
Figure 3: RNA-sequencing analysis of ARPE-19 cells treated with ROCK inhibitor Y-27632. A) Robin interprets results from the first experiment and proposes follow-up assays. B-D) Example plots from a Finch RNA-seq analysis, formatted for readability in publication by a human. B) Finch-made volcano plot showing differentially expressed genes between Y27632-treated and wildtype cells after phagocytosis. C) Finch-made consensus findings from eight RNA-seq analysis trajectories, showing the percentage of analyses that identified the same genes as consistently up- or down-regulated. D) Finch-made GO-term enrichment of differentially expressed genes.
Crucially, the DGE analysis identified a significant upregulation of ABCA1, a critical lipid efflux pump. The sources note that ABCA1 is essential for healthy RPE function and is in the same transporter family as ABCA4, a known target in macular degeneration. This mechanistic insight, generated through Robin's proposed experiment and Finch's analysis, highlights how the system can uncover novel molecular targets.
Further iterative refinement:
- Following the initial analysis, Robin generated a subsequent list of candidate drugs based on the insights gained. Human researchers tested 10 of these drugs, and Finch analyzed the data.
- Finch's analysis revealed that ripasudil, also a ROCK inhibitor and approved for glaucoma treatment in Japan, significantly enhanced RPE cell phagocytosis, performing even better than Y-27632 in this assay (Figure 4B). Robin's proposal for ripasudil drew on insights from the previous experimental round (Figure 4A). While the magnitude of effect differed slightly between Finch's analysis (7.5-fold increase) and human analysis (1.75-fold increase), both indicated a significant positive effect compared to control.
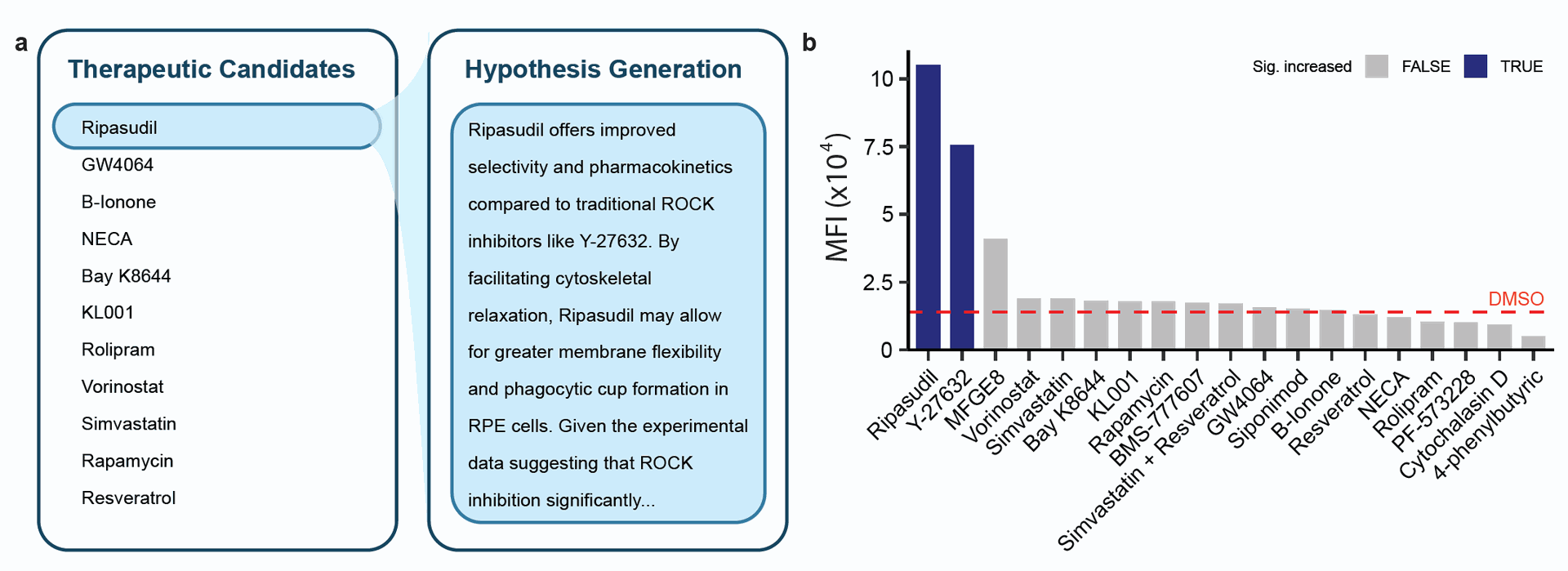
Figure 4: Ripasudil significantly enhances RPE phagocytosis. A) Excerpt of Robin proposal for ripasudil. Drawing from the insights from the first round of experimental analysis, Robin proposes ripasudil as a therapeutic candidate for treating dry AMD. B) Analyzed flow cytometry data from the second round of experiments shows that ripasudil significantly enhances phagocytosis in RPE cells, inducing an even greater effect than Y-27632.
This iterative process, where experimental results feed back into hypothesis generation, allowed Robin to progressively refine therapeutic candidates. Ripasudil's established safety profile for ocular use makes it a promising drug repurposing candidate for dAMD.
Conclusion
This research presents Robin, a pioneering multi-agent system that successfully integrates automated hypothesis generation and experimental data analysis, representing a new paradigm for AI-driven scientific discovery.
By applying Robin to dry age-related macular degeneration (dAMD), the system not only proposed enhancing RPE cell phagocytosis as a therapeutic strategy but also identified ripasudil as a promising therapeutic candidate and elucidated potential molecular targets like ABCA1 through iterative experimentation and autonomous data analysis.
The demonstration of Robin discovering and validating a novel drug repurposing candidate within an iterative lab-in-the-loop framework highlights its potential to significantly accelerate therapeutic discovery.
By automating the laborious processes of literature synthesis, hypothesis formulation, and initial data interpretation, Robin can potentially streamline research efforts and uncover insights that might be missed by human scientists alone.
One could interpret that systems like Robin pave the way for a future of scientific research where AI acts as a highly capable "co-scientist," working alongside human experts to navigate the complexity of modern biological data.
While the sources acknowledge areas for improvement, such as generating more detailed experimental protocols and enhancing the autonomy of the data analysis agent, the core achievement of automating the full intellectual cycle is substantial.
My interpretation is that the successful application to dAMD suggests this framework could be generalized to tackle therapeutic challenges in many other diseases, and perhaps even extend to non-therapeutic areas of biological research where iterative cycles of hypothesis, experiment, and analysis are central.
The research signifies a tangible step towards leveraging AI to accelerate fundamental scientific understanding and translate it more quickly into real-world applications. It has yet to be peer reviewed so the results will ultimately need to be verified before the community can benefit.

AI is Automating Therapeutic Drug Discovery: The Robin System by FutureHouse
Robin: A Multi-Agent System for Automating Scientific Discovery