Common infections become deadly because the bacteria causing them have learned to resist our best medicines. This is the growing crisis of antibiotic resistance. Scientists are urgently looking for new ways to fight these "superbugs," and a recent discovery offers a promising new approach
This NIH funded research from McMaster University and the University of Illinois identifies and characterizes lariocidin (LAR), a novel lasso peptide antibiotic produced by Paenibacillus sp. M2, which represents the first known member of this class of natural products to inhibit bacterial growth. The mechanism of action is believed to be the targeting of the ribosome and interfering with protein synthesis.
The study elucidates LAR's mechanism of action, its broad-spectrum antibacterial activity, its low propensity for resistance development, and its in vivo efficacy against a multidrug-resistant bacterial infection, highlighting a new avenue for antibiotic discovery.
Key Takeaways:
- Lariocidin (LAR) exhibits broad-spectrum antibacterial activity against Gram-positive, Gram-negative bacteria (including multidrug-resistant strains), and mycobacteria, as detailed in Table 1.
- LAR and its derivative lariocidin B (LAR-B) are the first identified lasso peptides that function as ribosome inhibitors, a major class of antibacterial targets.
- LAR binds to a unique site on the small ribosomal subunit (30S), distinct from other known ribosome-targeting antibiotics, interacting with the 16S rRNA and aminoacyl-tRNA. This is visually represented in Figure 4, which shows LAR's binding site relative to other antibiotics.
- LAR inhibits bacterial growth by interfering with translocation during protein synthesis and inducing miscoding, as demonstrated through in vitro assays and a reporter gene assay. Figure 2 illustrates the inhibition of translocation and the induction of miscoding.
- LAR demonstrates a low propensity for generating spontaneous resistance and is unaffected by common resistance mechanisms. The study identified specific mutations in the 16S rRNA of E. coli that confer resistance, pinpointing the drug's target on the ribosome.
- LAR shows no significant human cell toxicity or hemolytic activity at relevant concentrations, suggesting potential for therapeutic development. Table 1 provides the IC50 and hemolysis data.
- LAR exhibits potent in vivo activity in a mouse model of Acinetobacter baumannii infection, significantly reducing bacterial burden and improving survival rates. Figure 5 shows the reduction in bacterial burden and the Kaplan-Meier survival curve.
- The biosynthetic gene cluster (lrc BGC) responsible for LAR production was identified and functionally validated through heterologous expression in Streptomyces lividans, providing a means for further study and potential optimization. Figure 1 outlines the lrc BGC and confirms heterologous production.
- Lariocidin B (LAR-B), featuring a unique second intramolecular ring, represents a new class (Class V) of lasso peptides. Figure 3 illustrates the structure of LAR-B with its second isopeptide bond.
Overview
The escalating crisis of antibiotic resistance necessitates the discovery of novel antibacterial agents with unique mechanisms of action. Ribosomally-synthesized and post-translationally modified peptides (RiPPs) represent a promising source of such compounds due to their structural diversity and potent biological activities.
Among RiPPs, lasso peptides (LPs) are characterized by a distinctive knotted three-dimensional structure, formed by an N-terminal macrolactam ring through which a C-terminal tail is threaded.
While several LPs exhibit antibacterial properties by targeting various bacterial components like proteases or RNA polymerase, none were previously known to inhibit the ribosome, a historically successful target for antibiotics.
This groundbreaking research reports the identification and comprehensive characterization of lariocidin (LAR) and its internally cyclized derivative, lariocidin B (LAR-B), as the first ribosome-targeting lasso peptides. Produced by Paenibacillus sp. M2, these peptides demonstrate broad-spectrum antibacterial activity against a range of bacterial pathogens, including clinically relevant multidrug-resistant strains.
The researchers employed a combination of genomic analysis, biochemical assays, structural biology (X-ray crystallography), and in vivo infection models to elucidate the mechanism of action and therapeutic potential of lariocidins.
The identification of the biosynthetic gene cluster (BGC) responsible for lariocidin production and its successful expression in a heterologous host (Streptomyces lividans) provided crucial tools for studying these novel antibiotics.
The detailed structural analysis of LAR and LAR-B bound to the Thermus thermophilus ribosome revealed a unique binding site on the small ribosomal subunit, distinct from those of other known ribosome inhibitors. This novel interaction disrupts the essential process of protein synthesis by inhibiting translocation and inducing errors in the genetic code, ultimately leading to bacterial cell death.
Why It’s Important
The discovery of lariocidin as the first ribosome-targeting lasso peptide is significant step forward in the development of novel antibacterial agents. It expands the known functional repertoire of lasso peptides, establishing them as a new class of natural products capable of interfering with bacterial translation, a central process for life.
Given the well-validated nature of the ribosome as an antibiotic target, this finding opens up a new chemical space for the development of much-needed antibacterial drugs. The unique binding site of LAR on the ribosome offers a potential advantage in combating antibiotic resistance across a wide variety of bacterial types.
The study also shows that LAR is mostly insensitive to mutations or enzymatic resistance mechanisms that confer high levels of resistance to other ribosome-targeting antibiotics. As resistance to existing antibiotics continues to spread, the identification of inhibitors acting on novel sites with unconventional structures is crucial for increasing human survival rates.
The broad-spectrum antibacterial activity of LAR, including its efficacy against notorious ESKAPE pathogens like multidrug-resistant Acinetobacter baumannii, and its promising in vivo activity in a mouse infection model, underscore its therapeutic potential.
The lack of significant cytotoxicity against human cells and low hemolytic activity further support its promise as a potential drug scaffold. While the identification of lrc-like biosynthetic gene clusters in diverse bacteria suggests that LAR may represent a broader family of ribosome-targeting lasso peptides, hinting at a wealth of undiscovered natural products with similar mechanisms and potentially varying antibacterial spectra or pharmacokinetic properties.
The unique structural feature of LAR-B, with its double lariat fold, might confer enhanced stability or other beneficial properties, making it an interesting lead for drug development.
This research not only provides a new antibiotic candidate but also uncovers a new mechanism and structural class of ribosome inhibitors, paving the way for future drug design and optimization efforts to address the global antibiotic resistance crisis.
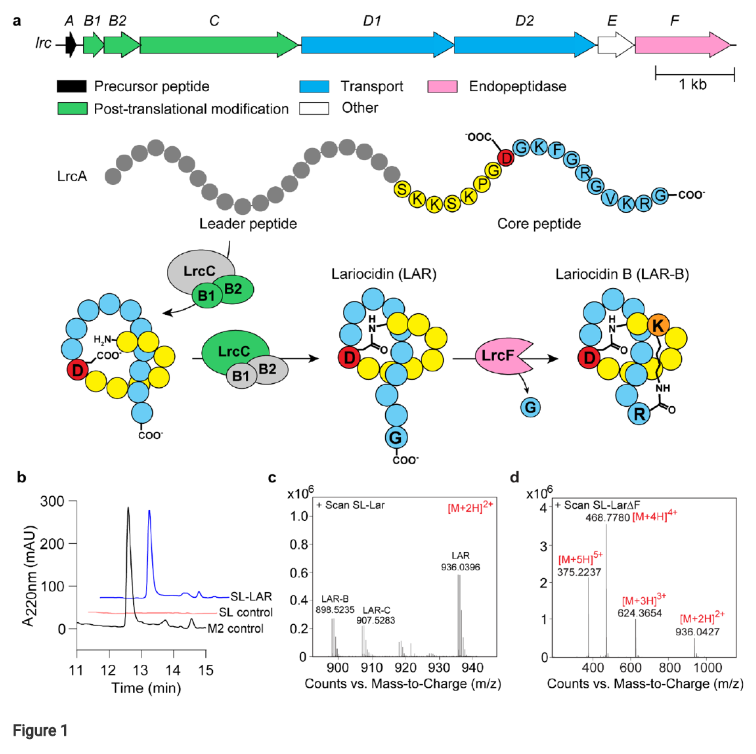
Figure 1. Lariocidin and its biosynthetic gene cluster. (a) Top, gene composition of the lrc BGC. Bottom, posttranslational modification of the LrcA precursor peptide leads to the production of LAR-A and LAR-B.(b) Heterologous expression of lrc BGC in Streptomyces lividans and analysis of LAR in cell-freesupernatant. The panel shows chromatographic analysis of LAR produced in the heterologous host. SL-Lar S. lividans pIJ10257-lrc; M2 control is LAR puried from the native producer Paenibacillus M2; SL-control is S. lividans with the empty vector (without lrc BGC). (c) LC-MS analysis of LAR purifed from theheterologous host. All masses shown are ions corresponding to [M+2H]2+. (d) LC-MS analysis of LAR.
Summary of Results
The researchers began by screening environmental bacterial isolates for antimicrobial activity, leading to the identification of Paenibacillus sp. M2, which produced an active compound against multidrug-resistant Acinetobacter baumannii and an antibiotic-hypersusceptible E. coli strain.
Bioactivity-guided purification and genomic analysis revealed that the active compound was a lasso peptide, named lariocidin (LAR), encoded by the lrc biosynthetic gene cluster (BGC).
Heterologous expression of the lrc BGC in Streptomyces lividans confirmed the identity of LAR as the product of this cluster (Figure 1). Chemical characterization revealed that LAR is an 18-amino acid peptide with a macrolactam ring, and two additional related compounds, LAR-B and LAR-C, were also identified. LAR-B was shown to possess a unique second intramolecular ring (Figure 3).
Antimicrobial testing demonstrated that LAR exhibits broad-spectrum bactericidal activity against various Gram-positive and Gram-negative bacteria, including multidrug-resistant clinical isolates (Table 1).
LAR did not cause cell lysis or membrane permeabilization, suggesting an intracellular target. Fluorescence microscopy showed that labeled LAR accumulates in the bacterial cytoplasm (Figure 2d).
Attempts to isolate spontaneous resistant mutants in E. coli and B. subtilis yielded mutations primarily in genes related to the electron transport chain, suggesting potential effects on uptake. However, using an E. coli strain with a single rRNA operon, the researchers successfully isolated LAR-resistant mutants with specific mutations in the 16S rRNA (Extended Data Fig. 6), implicating the ribosome as the target.
In vitro transcription-translation assays further confirmed that LAR inhibits protein synthesis with submicromolar potency, showing selectivity for bacterial over mammalian translation systems (Figure 2e-g).
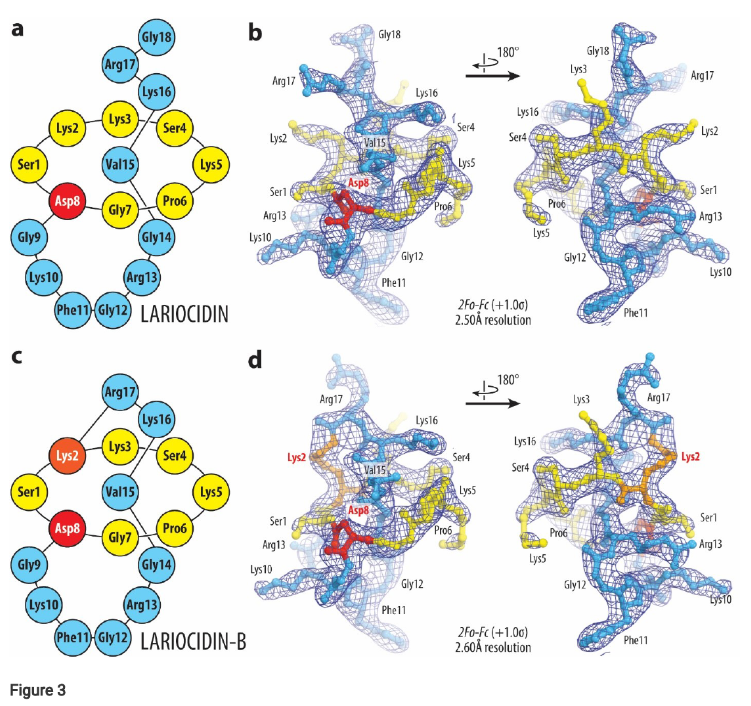
Figure 3. Structures and electron density maps of ribosome-boundLAR-A and LAR-B.(a, c) Schematic diagrams of the lasso peptides LAR-A (a) and LAR-B (c) highlighting their N-terminal residues 1-7 (yellow), branching point at residue 8 (red), and C-terminal residues 9-18 (blue). Lys2 residues of LAR-B forming the second isopeptide bond is colored orange. (b, d) 2 Fo-Fc Fourier electron density maps of LAR (b) and LAR-B (d) in complex with the T. thermophilus 70S ribosome (blue mesh). The rened models of lariocidins are displayed in their respective electron density maps after the renements contoured at 1.0σ. Color scheme as in panels (a) and (c), respectively.
To elucidate the binding mechanism, X-ray crystal structures of LAR and LAR-B in complex with the Thermus thermophilus ribosome were determined (Figure 3 and 4). These structures revealed that lariocidins bind to a unique site on the small ribosomal subunit, adjacent to the decoding center, interacting with helices h31, h32, and h34 of the 16S rRNA and the A-site tRNA.
This binding site is distinct from those of other known ribosome-targeting antibiotics (Figure 4e, f). Functional assays demonstrated that LAR inhibits ribosome translocation (Figure 2h) and induces miscoding (Figure 2j).
Finally, in vivo studies using a neutropenic mouse model of Acinetobacter baumannii infection showed that LAR significantly reduced bacterial burden in multiple tissues and improved survival rates (Figure 5).
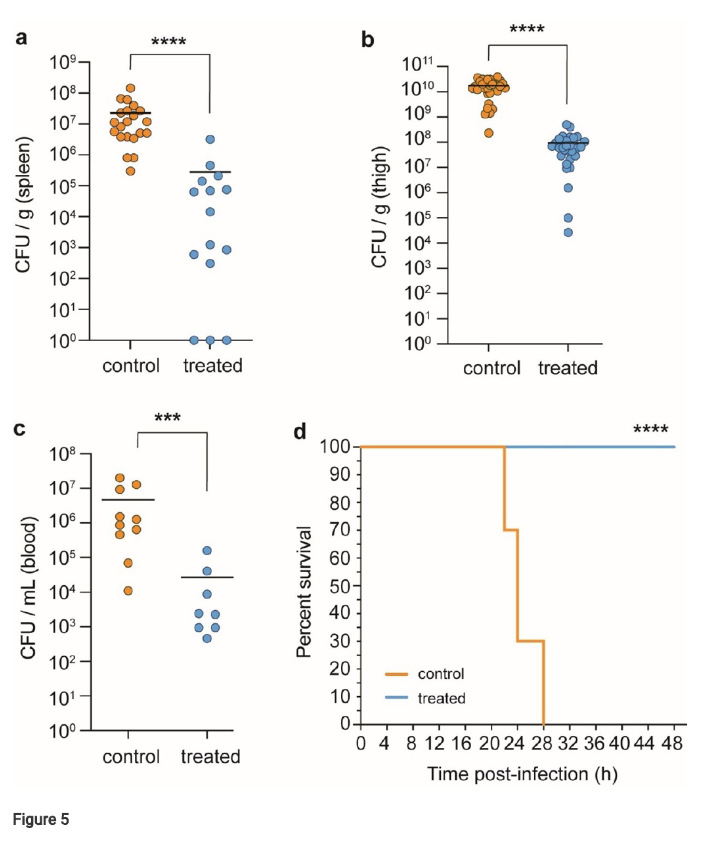
Figure 5. Therapeutic efficacy of Lar in a mouse neutropenic thigh infection model. (a-c) Reduction in bacterial burden (A. baumanniiC0286) after 24 hr post administration of LAR as measured by colony forming units (cfu) per gram of tissue or per mL of blood. (a)Bacterial burden in the spleen. control (n = 21) and treated (n = 15) groups. (b)Thigh bacterial burden in control (n = 32) and treated (n = 30) groups. (c)Blood bacterial burden in control (n = 10) and treated (n = 8) groups. Data points are from individual animals and horizontal lines represent the group means. Signicance was determined with a two-tailed Mann-Whitney test (*P≤0.05; **P<0.01; ***P<0.001;****P<0.0001). (d) Kaplan-Meier test showing group survival across select time points throughout A. baumannii thigh infection in vehicle control and LAR-treated mice. ****P <0.0001, Log-rank (Mantel-Cox) test.
Conclusion
This research successfully identified and characterized lariocidin (LAR) as the first ribosome-targeting lasso peptide antibiotic. Through a multidisciplinary approach encompassing genomics, biochemistry, structural biology, and in vivo pharmacology, the study elucidated LAR's novel mechanism of action, its broad-spectrum antibacterial activity against clinically relevant pathogens, its low potential for resistance, and its in vivo efficacy.
The discovery of this new class of ribosome inhibitors, exemplified by LAR and its unique binding site, offers a promising avenue for the development of alternative antibacterial therapeutics to combat the growing threat of antibiotic resistance. The structural novelty of LAR-B further expands the structural diversity of lasso peptides and suggests potential for further exploration and optimization of this class of natural products for medical applications.

A New Weapon Against Superbugs: Scientists Discover Lariocidin, a Lasso Peptide That Stops Bacteria in Their Tracks
A Broad Spectrum Lasso Peptide Antibiotic Targeting the Bacterial Ribosome