Imagine a constant battle happening inside our bodies, an evolutionary arms race between invading bacteria and the immune cells that protect us. Some bacteria, like Salmonella enterica serovar Typhimurium (the specific type often linked to food poisoning, which we'll call Salmonella here), are particularly tricky.
They don't just fight from the outside; they invade our own immune cells, specifically ones called macrophages. Macrophages are like the body's security guards and cleanup crew, designed to find, engulf, and destroy harmful microbes like bacteria.
However, Salmonella has developed clever strategies not just to survive inside these macrophages, but to actually use them as a safe place to multiply and hide.
This ability to survive the macrophage's attack is a key reason Salmonella can cause disease, ranging from stomach upset to life-threatening infections, especially in people with weakened immune systems. With the rise of antibiotic-resistant Salmonella strains, finding new ways to fight these bacteria is becoming increasingly urgent.
Understanding exactly how Salmonella pulls off this survival act inside our cells is crucial for developing new treatments.
This research dives deep into Salmonella's genetic playbook during macrophage infection. By looking at which bacterial genes are active at different times, researchers have uncovered a critical defense mechanism: the phage shock protein (Psp) system. This system helps Salmonella defend against natural antibiotic molecules produced by our immune cells, offering new clues about how these bacteria win the battle within.
Key Takeaways
- This study provides a comprehensive temporal analysis of the S. Typhimurium transcriptome during the course of macrophage infection, identifying key stages of bacterial adaptation.
- The phage shock protein (Psp) system is shown to be highly upregulated throughout macrophage infection, suggesting a critical role in intracellular survival.
- The research provides the first direct evidence that the Psp system in S. Typhimurium mediates resistance to cationic antimicrobial peptides (cAMPs), specifically CRAMP (cathelin-related antimicrobial peptide), within macrophages.
- The study surprisingly reveals that the SsrA-SsrB two-component system (TCS), traditionally known for regulating virulence genes within the Salmonella pathogenicity island 2 (SPI-2), plays a role in regulating the psp operon. This finding challenges the previous understanding of the regulatory network controlling antimicrobial resistance in Salmonella.
- The findings highlight the intricate interplay between bacterial stress responses and host defenses, offering potential new targets for anti-Salmonella therapies.
Overview
When Salmonella gets engulfed by a macrophage, it gets trapped inside a bubble-like compartment called the Salmonella-containing vacuole, or SCV. While this SCV offers the bacteria some protection and access to nutrients, it's also a dangerous place.
The macrophage tries to destroy the bacteria within the SCV using various weapons: acid, limiting essential nutrients, and deploying toxic molecules like natural antibiotics (called antimicrobial peptides or AMPs), reactive oxygen species (ROS), and reactive nitrogen species (RNS).
To survive this onslaught, Salmonella relies on sophisticated genetic programs that allow it to sense the dangers in its environment and quickly adapt its defenses.
Previous studies gave us glimpses into these programs, but newer technology allows for a much more detailed look.
This research used advanced RNA-sequencing (RNA-seq) to track Salmonella's gene activity at four distinct stages of infection (onset, early, middle, and late) inside primary mouse macrophages – cells taken directly from bone marrow to better mimic a real infection. These stages represent the different challenges Salmonella faces as the macrophage steps up its attack over time.
Why it Matters
Figuring out Salmonella's survival tricks inside macrophages is key to developing better treatments. Current antibiotics are becoming less effective due to resistant strains, making the search for new ways to combat these infections a global health priority.
This study shines a spotlight on the Psp system. Researchers found it acts like a shield, protecting Salmonella from the host's attack, specifically against a potent antimicrobial peptide called CRAMP (the mouse version of a similar human peptide).
Discovering that the Psp system – previously thought to mainly manage energy and membrane stress – directly fights off a key immune weapon opens up exciting possibilities for new therapies. If we could develop drugs to disable the Psp system, we might make Salmonella vulnerable to our immune system or even make existing antibiotics effective again.
Researchers also found something unexpected: the Psp system is controlled, in part, by another known bacterial system called SsrA-SsrB.
The SsrA-SsrB system is like a master switch for many of Salmonella's invasion tools, particularly those needed to live and multiply inside the SCV. Finding this connection shows that Salmonella's defense mechanisms (like Psp) are tightly linked to its attack strategies (controlled by SsrA-SsrB).
This complex interplay highlights how sophisticated Salmonella's adaptation is. Targeting the SsrA-SsrB switch might therefore have wide-ranging effects, disrupting both Salmonella's ability to cause disease and its ability to withstand our defenses.
Summary of Results
To understand Salmonella's adaptation strategy, researchers tracked its gene activity at 0, 4, 8, and 12 hours after infecting macrophages.
- Rapid Genetic Changes: The bacteria drastically changed their gene activity soon after entering the macrophage. About half of Salmonella's genes showed altered activity levels during the infection compared to the very beginning. The patterns of gene activation and deactivation over time fell into four main groups, showing a dynamic response to the changing conditions inside the SCV. For instance, genes related to energy production were turned down as the infection progressed, suggesting the bacteria shift their metabolism (See Figure 1).
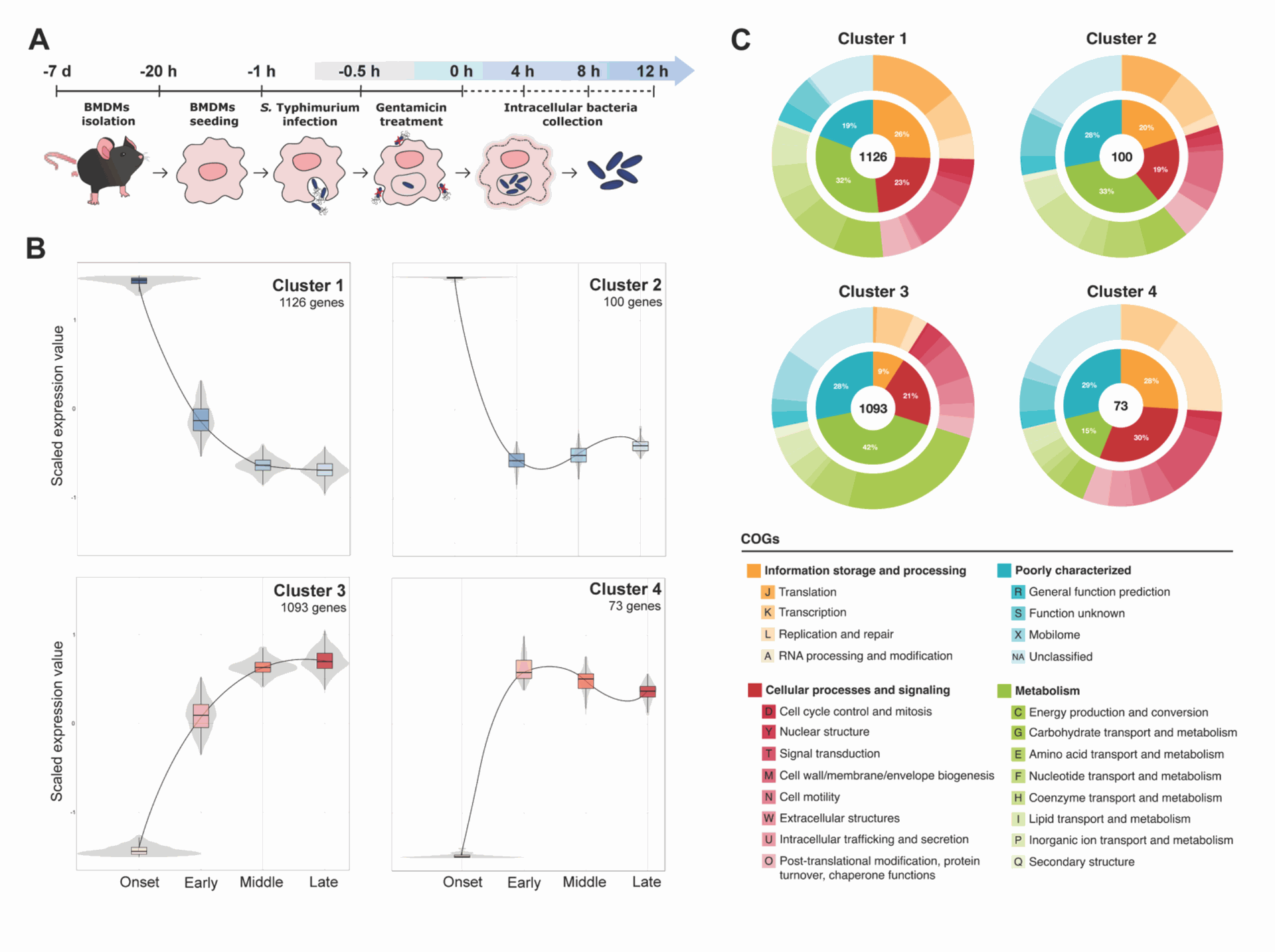
Figure 1 visually represents these findings. Part A shows the experiment's timeline. Part B groups genes based on when they were turned on or off during the infection. Part C shows the types of jobs these genes do, revealing which bacterial processes were most active at different stages.
- The Psp System: A Constant Defender: One defense system stood out: the phage shock protein (Psp) system. Genes belonging to this system were highly active throughout all stages of the infection, from early to late. The Psp system involves several genes (pspA through pspG) known to help bacteria cope with stress, especially stress that damages their protective outer layers. Its constant high activity suggested it plays a vital role in surviving the long haul inside the macrophage (See Figure 2).
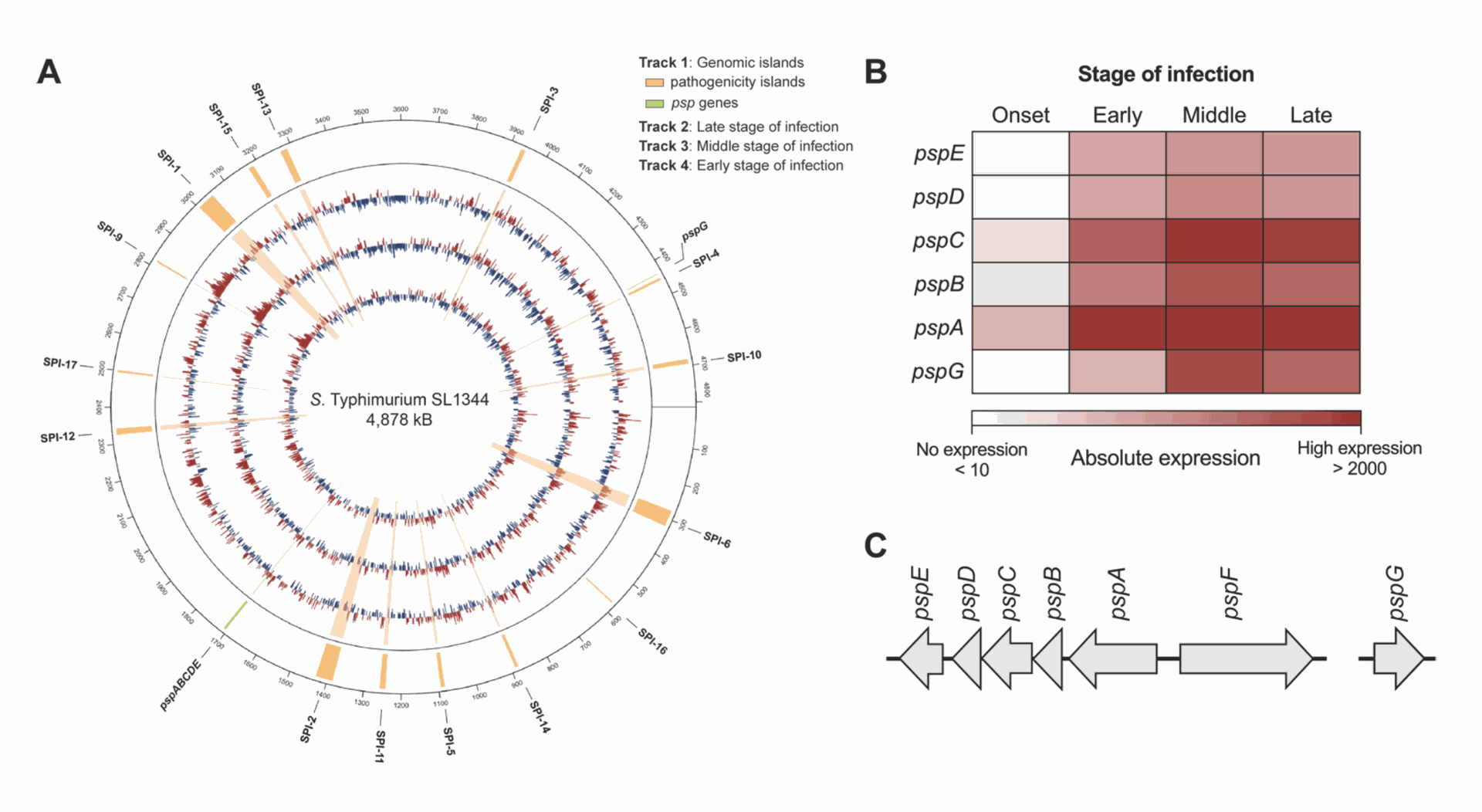
Figure 2 highlights the Psp system's activity. Part A is a circular map of the Salmonella genome showing gene activity changes; the tall red peaks in the Psp region indicate strong activation. Part B uses a heatmap (red intensity shows activity level) to confirm that individual Psp genes were highly active across all time points. Part C simply shows how the Psp genes are arranged on the bacterial chromosome.
- Psp vs. Antimicrobial Peptides: Since macrophages use antimicrobial peptides (AMPs) like CRAMP to kill bacteria, the researchers tested if the Psp system helps Salmonella resist these molecules. They found that Salmonella bacteria genetically engineered to lack a key Psp gene (pspA) were much more easily killed by CRAMP in lab tests. Importantly, these mutant bacteria also couldn't multiply properly inside normal macrophages. However, when these mutant bacteria infected macrophages that couldn't produce CRAMP (from genetically modified mice), they survived and multiplied much better. This was strong evidence that the Psp system directly protects Salmonella against CRAMP inside macrophages (See Figure 4).
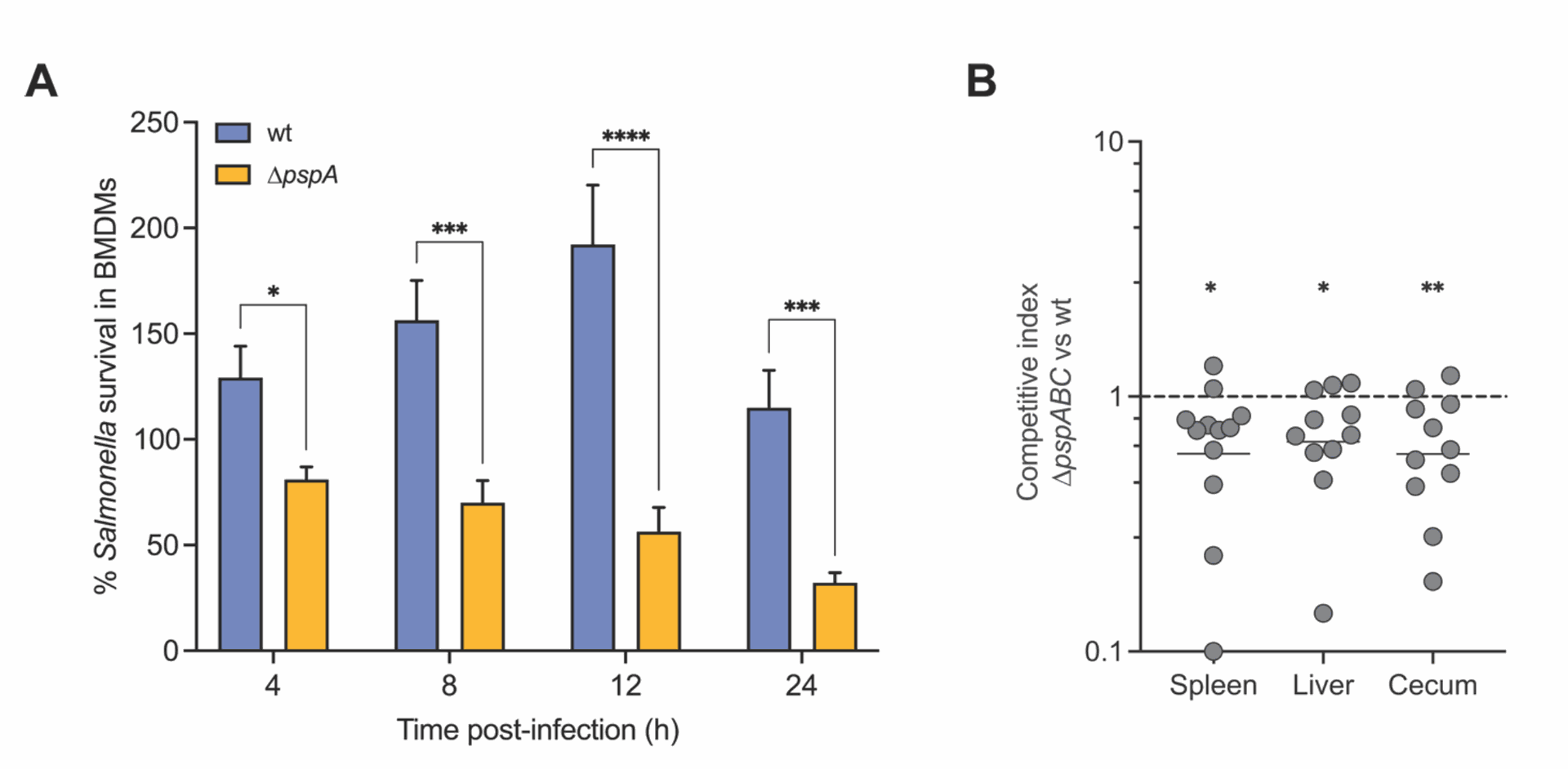
Figure 4 provides key evidence. Part A shows that Salmonella without the pspA gene (∆pspA) dies much more readily when exposed to CRAMP compared to normal (wt) Salmonella. Part D shows that while the ∆pspA mutant struggles to replicate inside normal macrophages (BMDMs), this defect disappears in macrophages that lack CRAMP (Camp-/-). This directly links PspA to resisting CRAMP within the macrophage.
- An Unexpected Controller: SsrA-SsrB: How does Salmonella know when to switch on the Psp defense? While other systems (PhoP-PhoQ, PmrA-PmrB) are known to control resistance to AMPs, they didn't seem to control the Psp genes. Surprisingly, the study found that the SsrA-SsrB system, primarily known for activating invasion genes, also activates the Psp system. Experiments showed that the SsrB protein directly turns on the main group of Psp genes (pspABCDE) by binding to a specific spot on the bacterial DNA near the pspA gene. This discovery links a major invasion control system directly to a key stress defense system.
Conclusion
This research provides a detailed timeline of how Salmonella adjusts its genes to survive inside our immune cells. They've identified a critical role for the phage shock protein (Psp) system in shielding Salmonella from the host's antimicrobial peptides, specifically CRAMP.
The unexpected discovery that the major invasion regulator SsrA-SsrB also controls this Psp defense reveals a new layer of complexity in Salmonella's survival strategy.
These findings deepen our understanding of the intricate balance Salmonella strikes between hiding from the immune system and adapting to the harsh environment within macrophages.
By learning more about how the Psp system works and how it's controlled, we can hopefully develop novel therapies that disarm these bacterial defenses. Understanding and disrupting these key survival mechanisms offers a promising path forward in the fight against drug-resistant Salmonella infections.
The Battle Within: How Salmonella Bacteria Survive Inside Our Immune Cells
The Salmonella phage shock protein system is required for defense against host antimicrobial peptides